eHypertension: A prospective longitudinal multi-omics essential hypertension cohort
- 18 April 2022
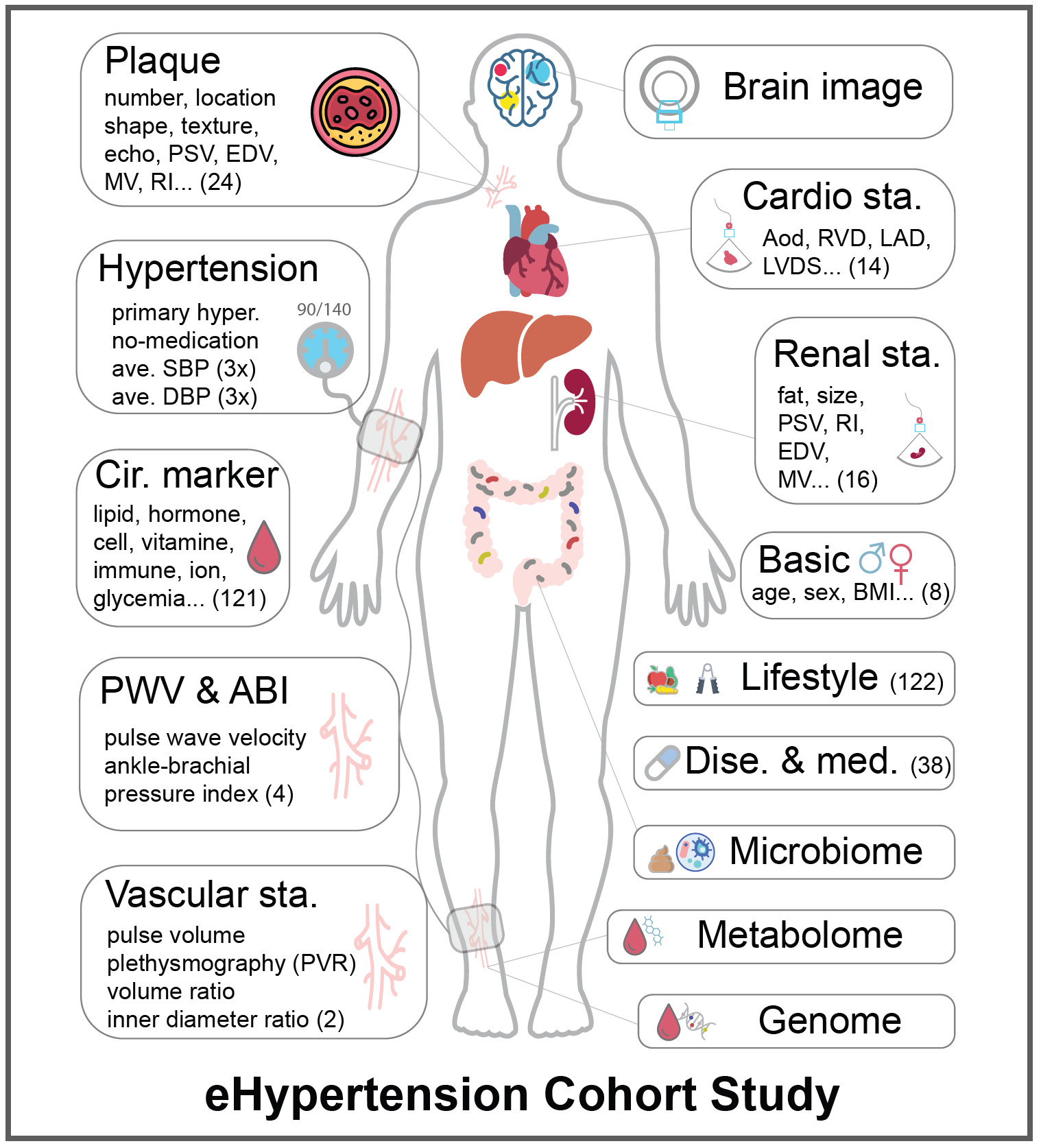
eHypertension is an essential hypertension cohort with multiple layers of omics data collected and will be followed up longitudinally. Randomized antihypertensive intervention has been set up in eHypertension. eHypertension is a unique cohort for studying the etiology and clinical outcome of hypertension. Interventions in eHypertension may contribute to the development of prediction models for personalized antihypertensive treatment.
Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses
- 16 March 2022

Majorbio cloud platform, which is a one-stop, online analytic platform for high-throughput omics data. The platform consists of three modules, which are pre-configured bioinformatic pipelines, cloud toolsets, and online omics’ courses. The Majorbio cloud platform was released at 26/10/2016, and ever since has been used by 70,000+ researchers from 3,562 institutes. It demonstrates that a one-stop, comprehensive online platform can facilitate the use of multi-omics data in all fields of biological analysis.
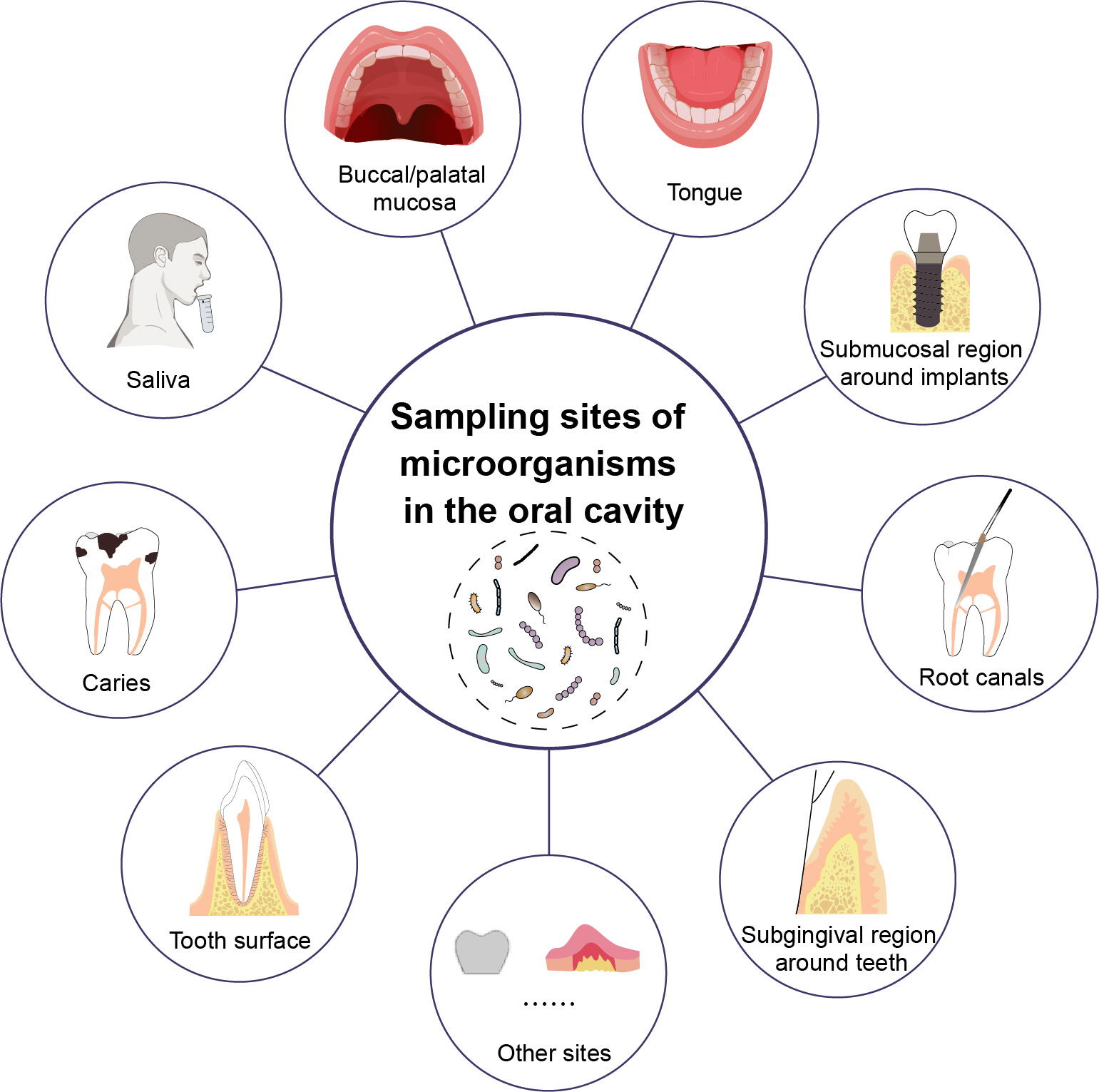
Standardized studies of the oral microbiome: From technology-driven to hypothesis-driven
- 11 April 2022
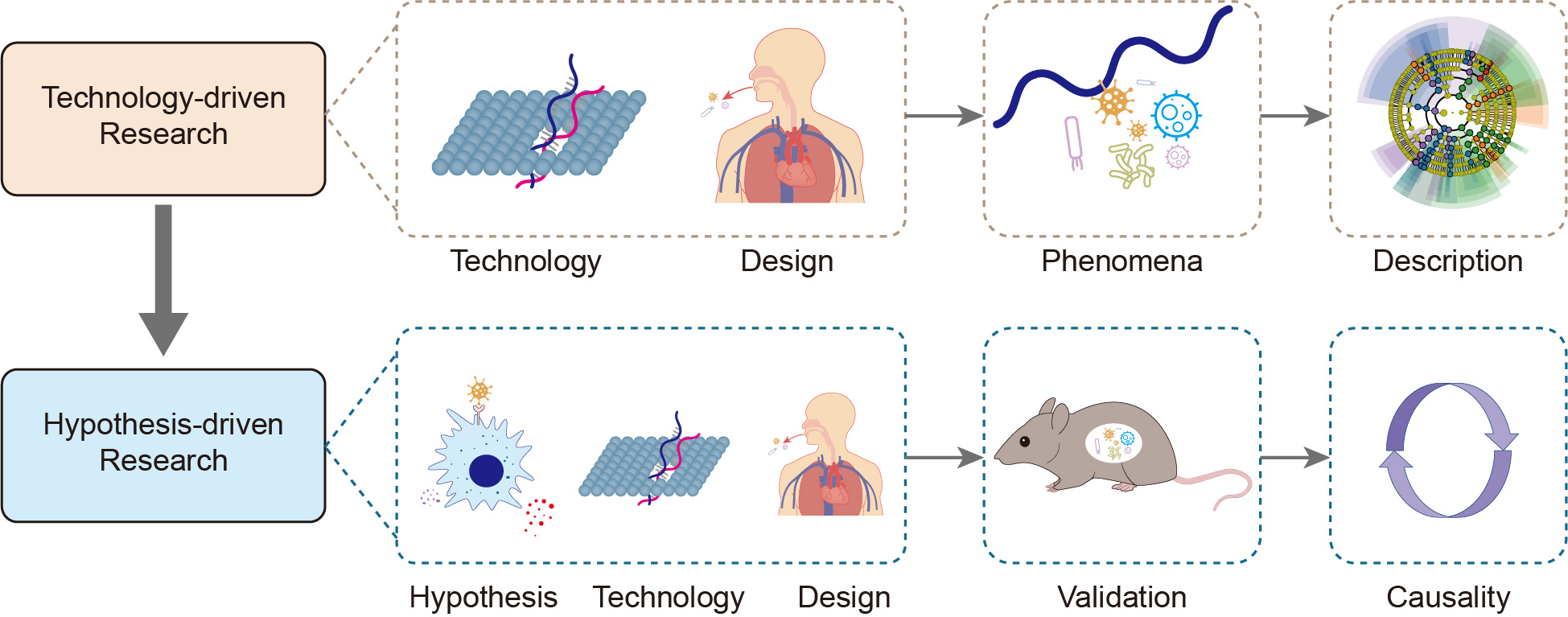
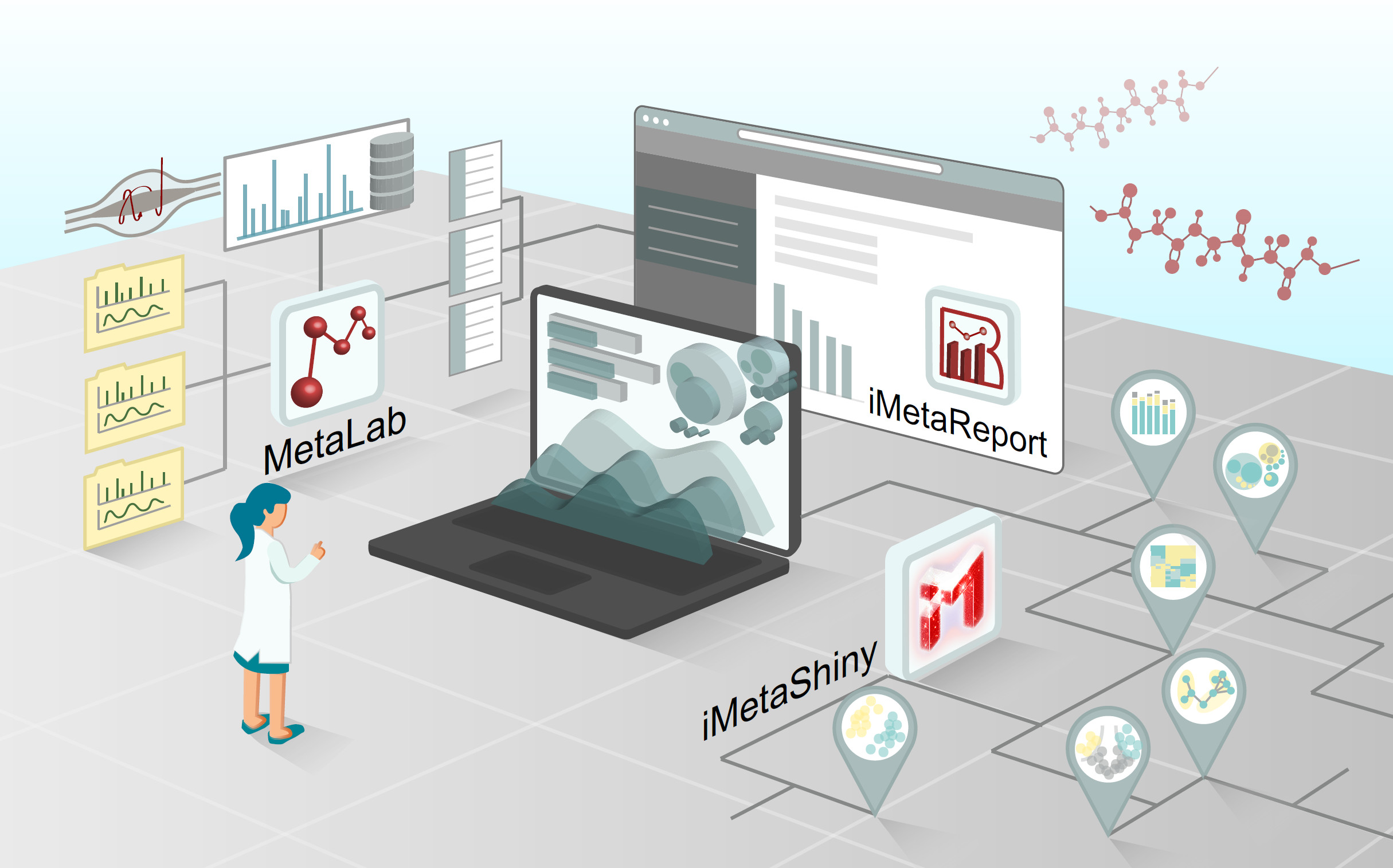
The microbiome is in a symbiotic relationship with the host. Among the microbial consortia in the human body, that in the oral cavity is complex. Instead of repeatedly confirming biomarkers of oral and systemic diseases, recent studies have focused on a unified clinical diagnostic standard in microbiology that reduces the heterogeneity caused by individual discrepancies. Research has also been conducted on other topics of greater clinical importance, including bacterial pathogenesis, and the effects of drugs and treatments. In this review, we divide existing research into technology-driven and hypothesis-driven, according to whether there is a clear research hypothesis. This classification allows the demonstration of shifts in the direction of oral microbiology research. Based on the shifts, we suggested that establishing clear hypotheses may be the solution to major research challenges.
iNAP: An integrated network analysis pipeline for microbiome studies
- 16 March 2022
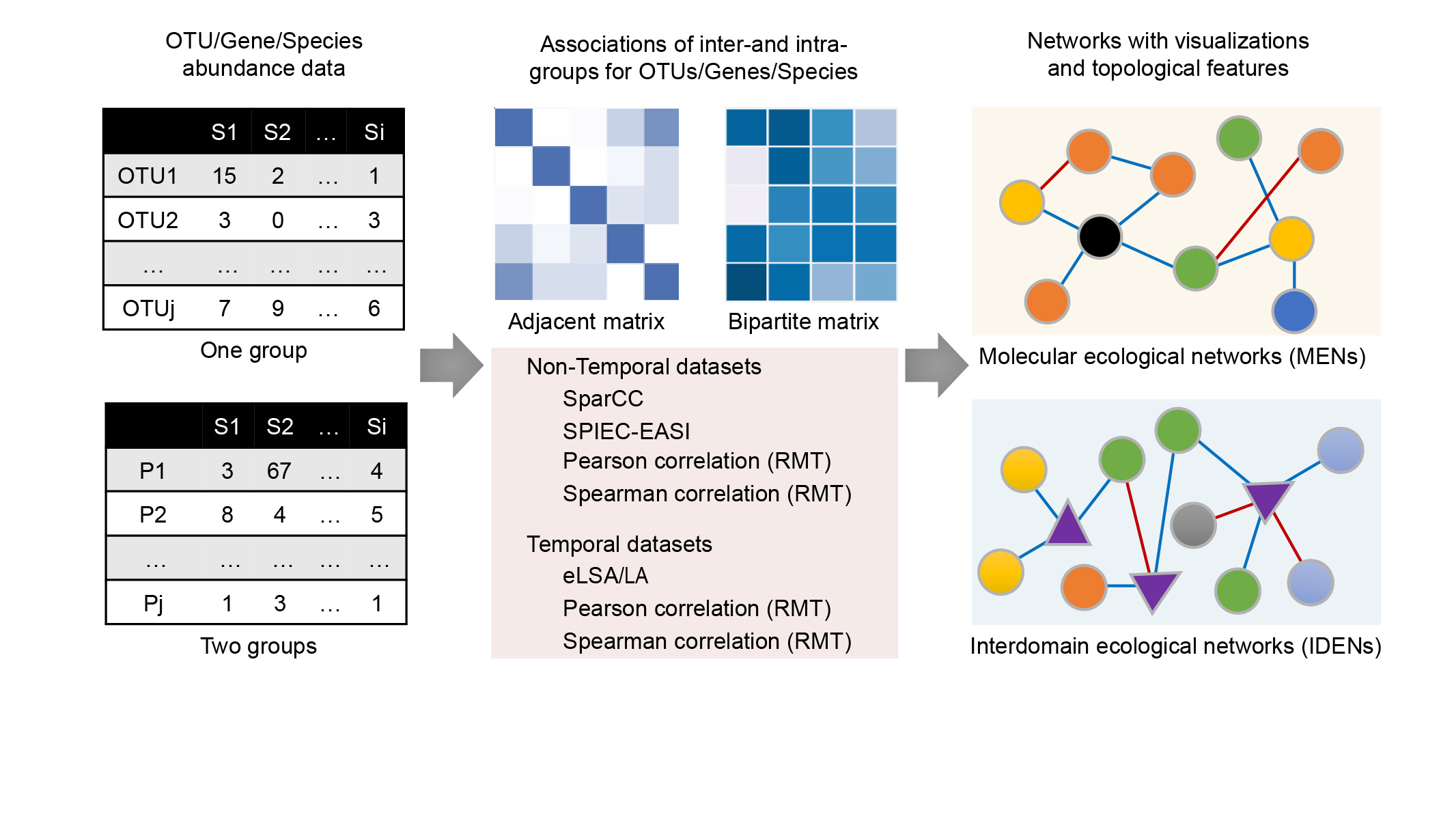
Integrated network analysis pipeline (iNAP) contains two network analyses pipelines for intradomain and interdomain associations of microbial species at multiple taxonomic levels, that is, molecular ecological network analysis pipeline and interdomain ecological network analysis pipeline. iNAP provides multiple approaches and methods for network analysis of microbiome studies, including SparCC, eLSA, SPIEC-EASI, and RMT-based Pearson's or Spearman's correlations.
The impact of aquaculture system on the microbiome and gut metabolome of juvenile Chinese softshell turtle (Pelodiscus sinensis)
- 5 April 2022
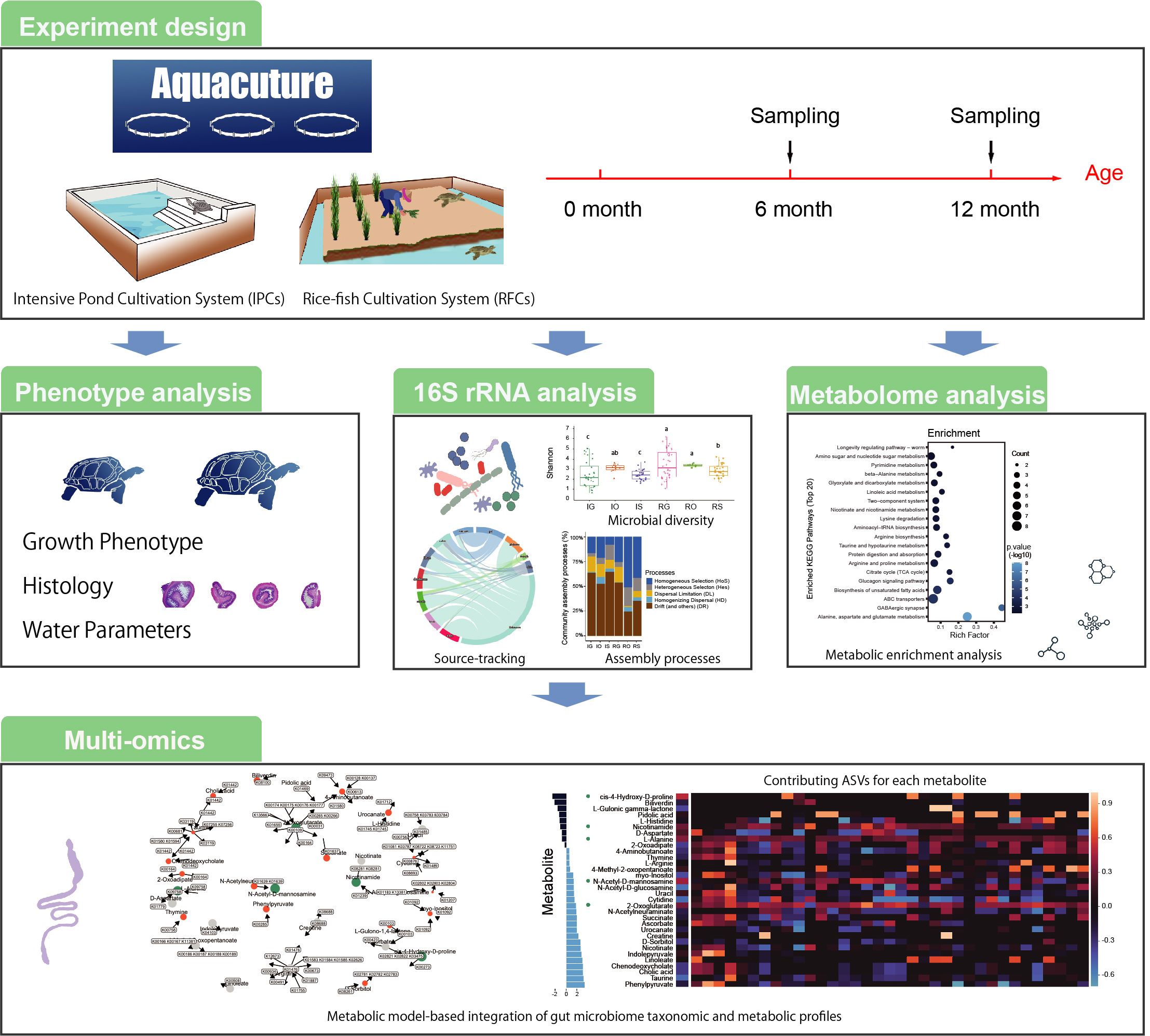
Different aquaculture system induces distinct phenotype, microbiome assembly, and gut metabolome. In brief, different aquaculture system induces distinct tissue-specific microbiome assembly that alters host fitness in part through gut metabolites. Distinct microbial diversity and community structure are driven through different sources and community assembly processes. To identify several key genes and microbial species that contribute to five significantly changed gut metabolites.
Differential microbial assembly processes and co-occurrence networks in the soil-root continuum along an environmental gradient
- 5 April 2022
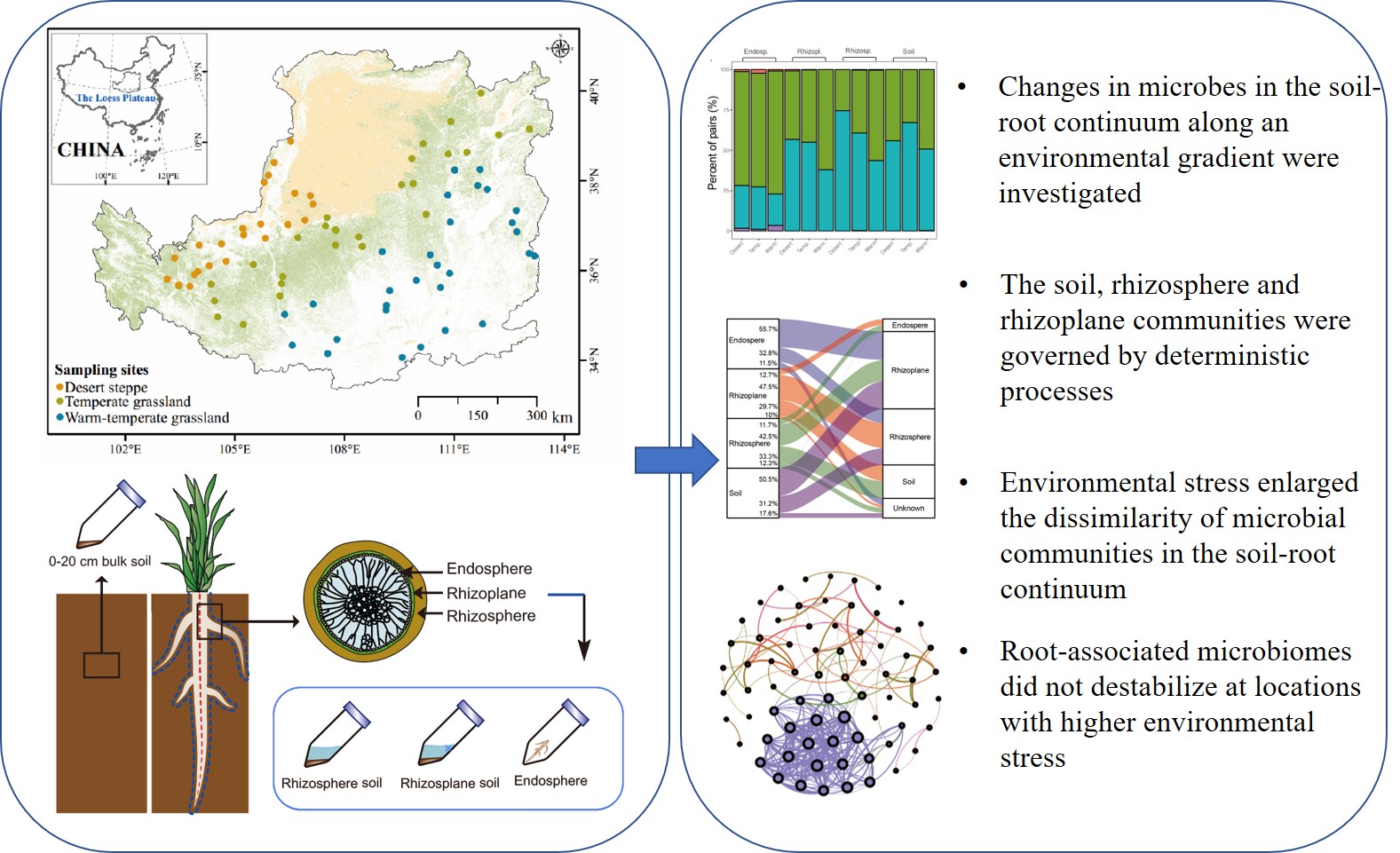
Changes in microbes in the soil-root continuum along an environmental gradient were investigated. The soil, rhizosphere and rhizoplane communities were governed by deterministic processes. Environmental stress enlarged the dissimilarity of microbial communities in the soil-root continuum. Root-associated microbiomes did not destabilize at locations with higher environmental stress.
A neural network-based framework to understand the type 2 diabetes-related alteration of the human gut microbiome
- 5 May 2022
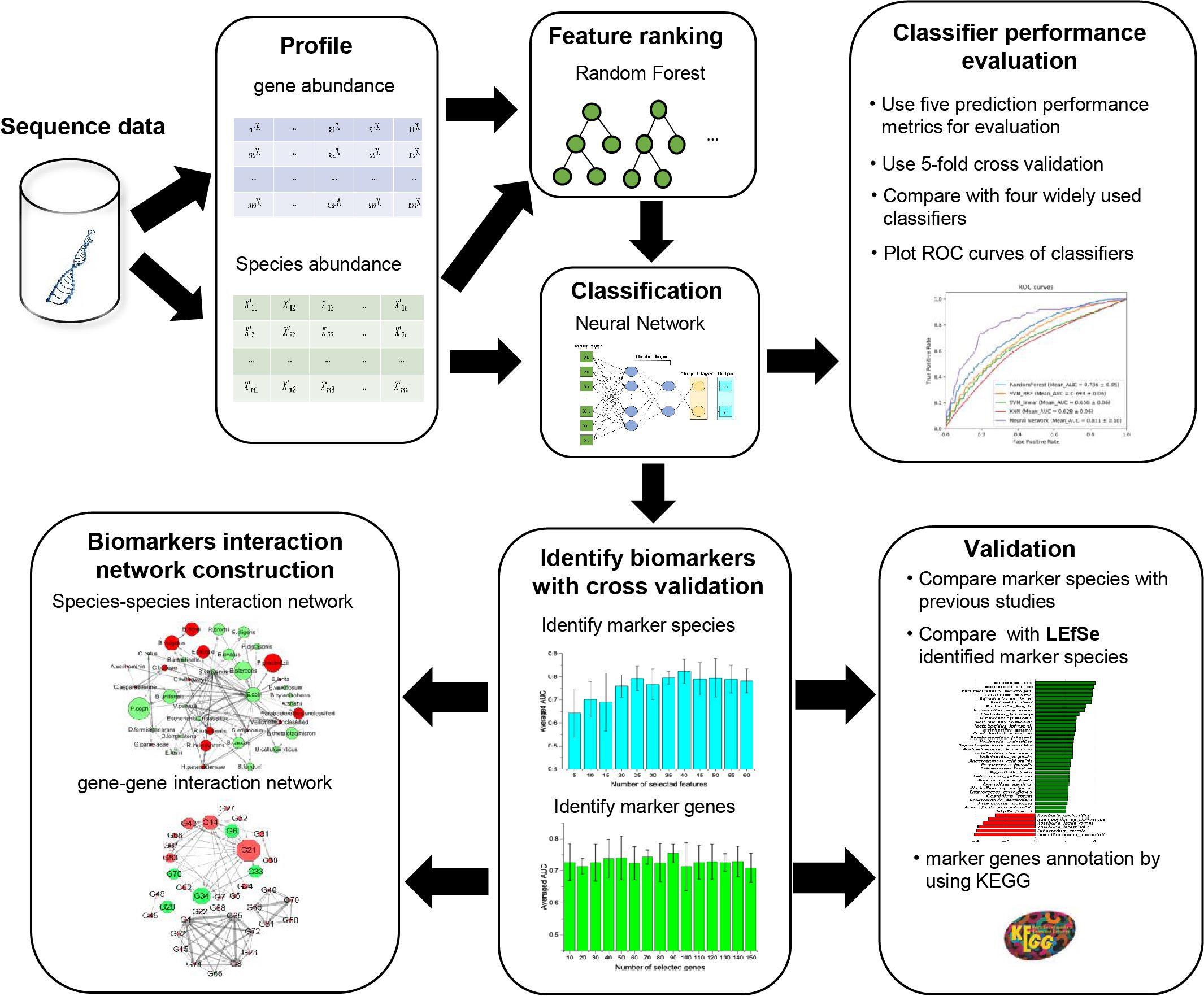
A framework combining neural network and random forest for identifying the type 2 diabetes (T2D)-related biomarkers. Constructing the directed interaction networks of the biomarkers for analyzing the potential drivers of the microbial community associated with T2D. Analyzing the covary of the biomarkers with the dynamic change of fasting blood glucose in the development of T2D.
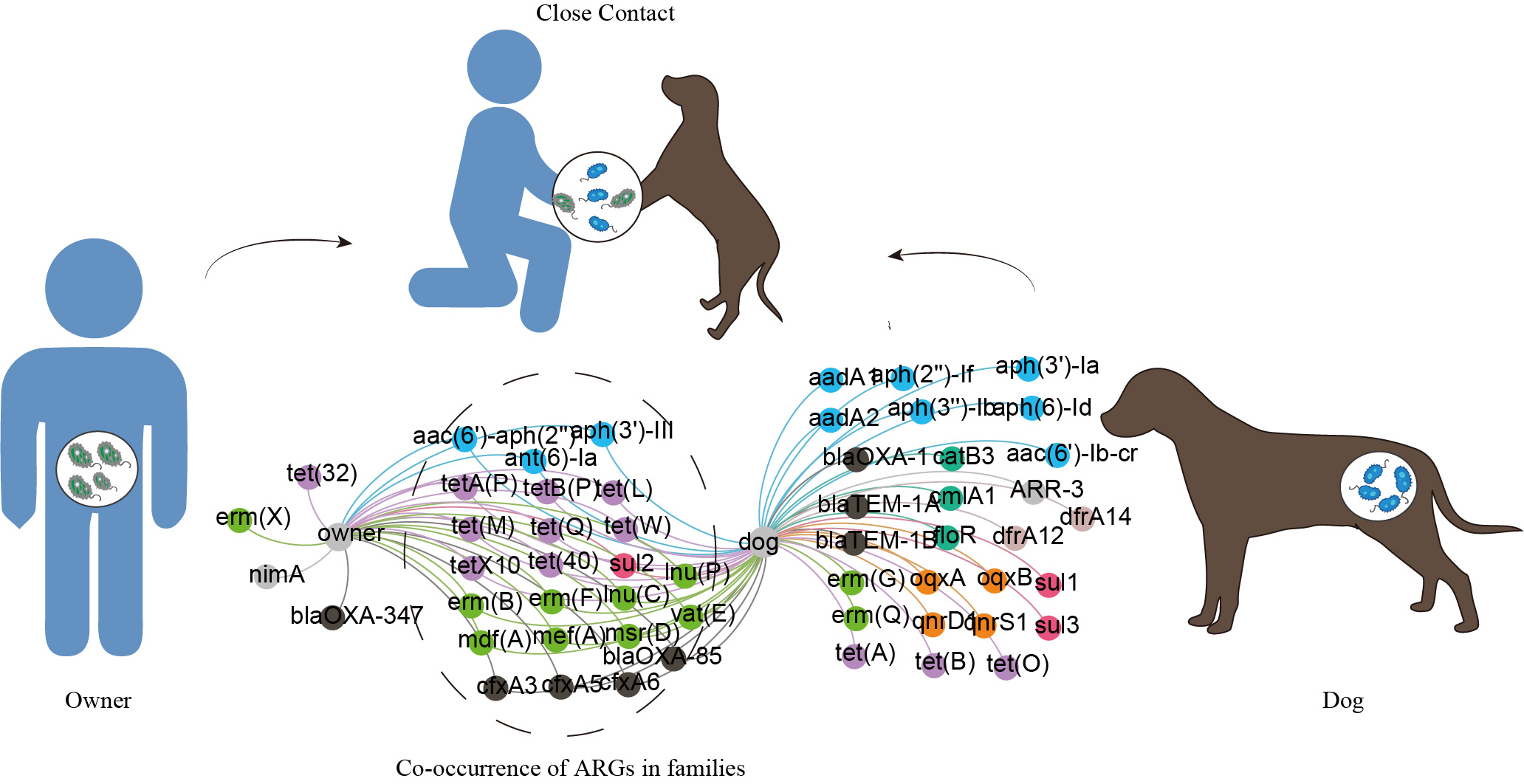
The co-occurrence of antibiotic resistance genes between dogs and their owners in families
- 04 May 2022
Microorganisms as bio-filters to mitigate greenhouse gas emissions from high-altitude permafrost revealed by nanopore-based metagenomics
- 08 May 2022
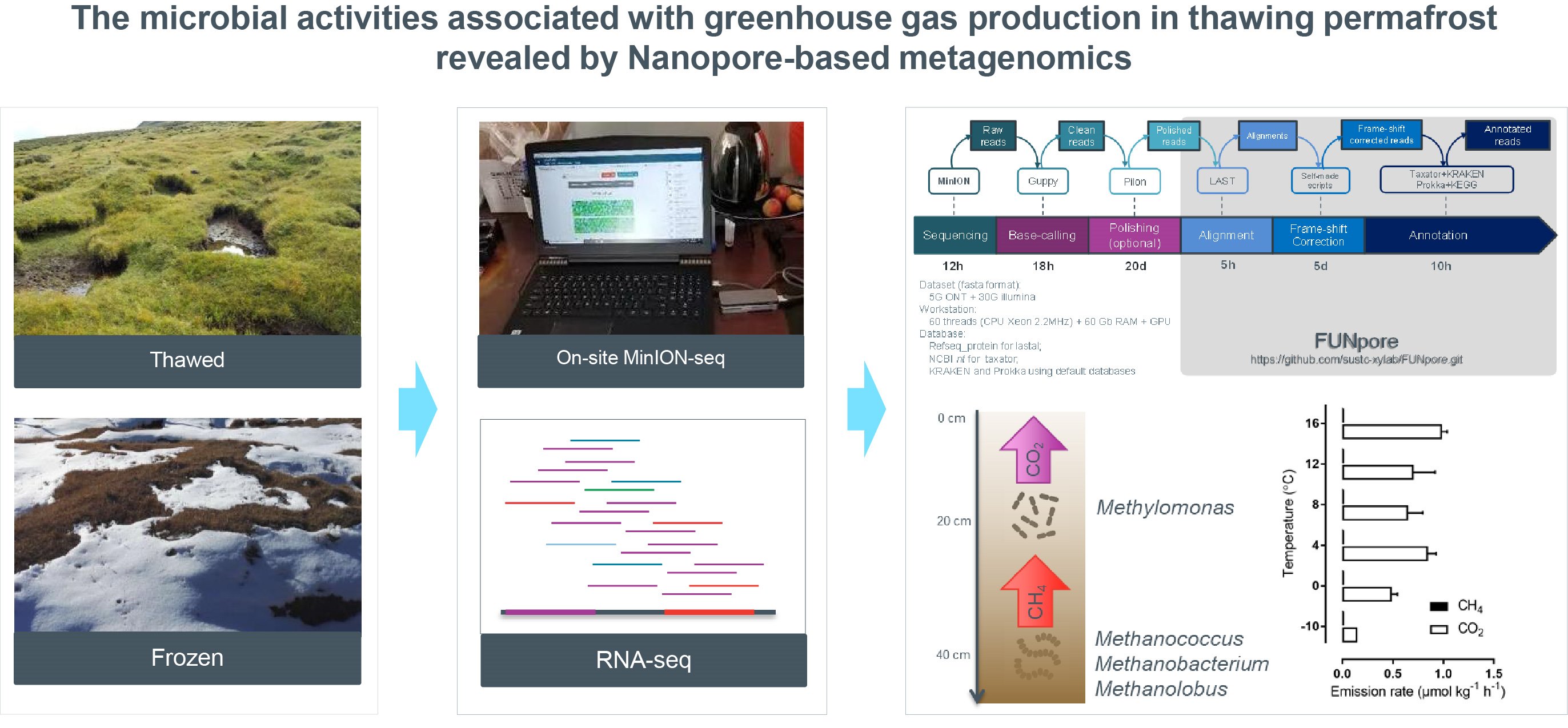
On-site MinION metagenomics and RNA-seq were combined to explore microbial functionalities of active layers of the soil of Qilian Mountain permafrost. With the developed annotation pipeline-FUNpore, 42% more genera and 58% more genes were detected. Active methane consumption by Methylomonas could serve as a biofilter to mitigate CH4 emission from permafrost. Dissimilatory nitrate reduction to ammonia pathway enabled a closed microbial nitrogen cycle and N2O consumption was observed.