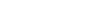Artificial intelligence-driven anticancer peptide discovery
- 05 November 2025
This review systematically summarized 68 artificial intelligence (AI) models for anticancer peptide (ACP) screening and presented a comprehensive AI-based framework. The framework consists of six components: data collection and organization, feature extraction, model construction, interpretability analysis, experimental validation, and integration with multi-omics and biotechnologies. This strategy improves screening efficiency and accuracy, addresses the limitations of interpretability and wet-lab validation, and accelerates the translation of AI-discovered ACPs into clinical applications.
Microbiota-gut-brain axis multi-organ chip construction and applications in drug evaluation
- 24 November 2025

Microbiota-gut-brain axis multiorgan chip and its applications in drug evaluation. The chip includes essential biological components such as gut microbiota, gut barrier, blood-brain barrier (BBB), and brain. This integrated platform incorporates neural (blue), endocrine (yellow), immune (pink), and metabolic (green) pathways to investigate the impact of drugs on neural function, BBB permeability, microbiota–host interactions, and gut barrier function. Furthermore, the platform is utilized for mechanism analysis of cross-organ interactions, drug screening, and preclinical drug research, such as pharmacokinetics, pharmacodynamics, and toxicity evaluation. It also supports the development of microbiota modulation strategies and provides innovative tools for targeted drug delivery for neurological diseases.
Species-level exploration of the gut microbiome in the leaf-eating Presbytis monkeys reflected the effects of anthropogenic activity and specialized dietary niches: Conservation on the fourth biodiversity level
- 16 September 2025
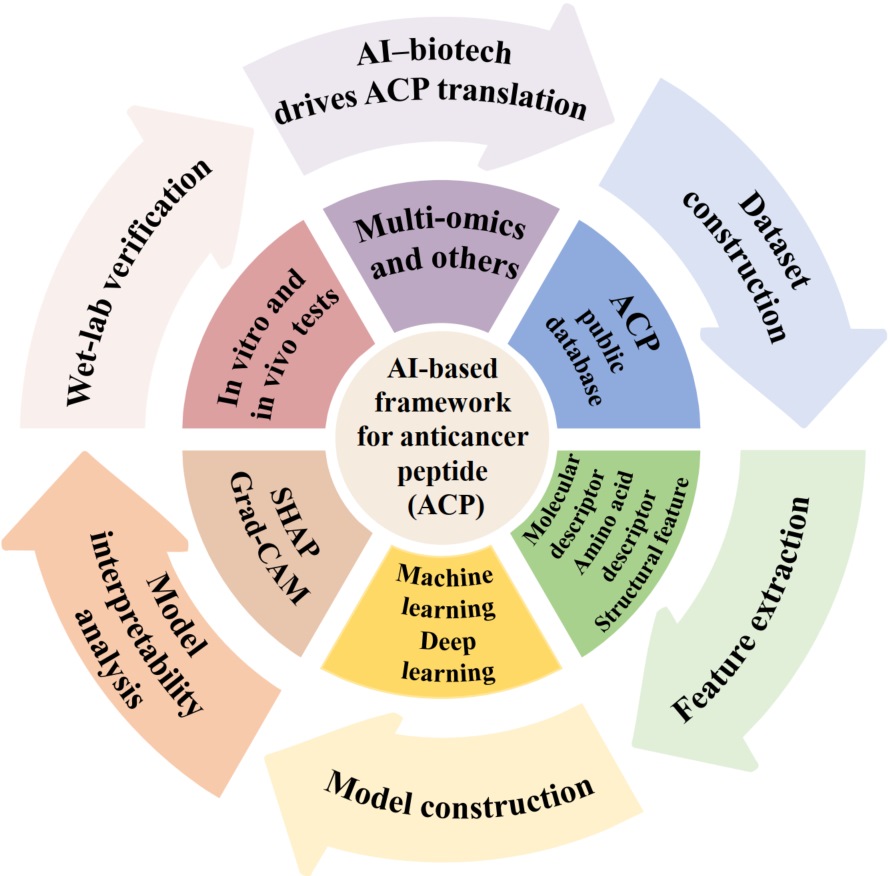
Seventy-five percent of nonhuman primates have declining populations caused by deforestation and habitat loss, mainly due to human disturbance worldwide. Endangered white-headed langurs and François' langurs are two allopatric species inhabiting limestone forests. Given the negative effects of human disturbance on the gut microbial community, we investigated the gut microbial composition and function in these two wild langurs under human disturbance (with the zoo as a reference for the strongest disturbance) using multiple methods (landscape, nutritional analysis, behavioral observation, 16S rRNA gene full-length sequencing of 237 samples, and deep metagenomic sequencing). We found that the relative abundance of four typical human disturbance-related (HDR) gut microbial families (Lachnospiraceae and Bacteroidaceae) increased with growing human disturbance. We identified the gut microbial species involved in the main dietary or host-derived component digestion (e.g., diet-derived polysaccharides, oxalate, and host-derived glycans) based on a comparative genomic analysis of the non-redundancy of 1199 metagenomic assembled genomes. These langurs harbored some putative common cellulolytic and oxalate-degrading bacteria, and also harbored some host-specific bacteria. Thus, we conclude that increased HDR species in the langur population might be caused by dietary changes and increased sugar intake, such as steamed bread of corn and sorghum, fruits in the zoo, and anthropogenic diet, corn, and sweet potatoes in wild langurs. Factors such as diet, host, and living environment under anthropogenic disturbance may influence the gut microbial community and function in these langurs. The langur gut microbiome plays a potential role in helping the host adapt to the leaf diet and limestone living environment. In the future, wild primate conservation should not only focus on host genetic diversity but also on the changes in symbiotic microbiomes under anthropogenic disturbance.
Cross-continental transmission and host adaptation of Mycobacterium tuberculosis in China unveiled by population history reconstruction and adaptive evolution signal detection
- 20 September 2025
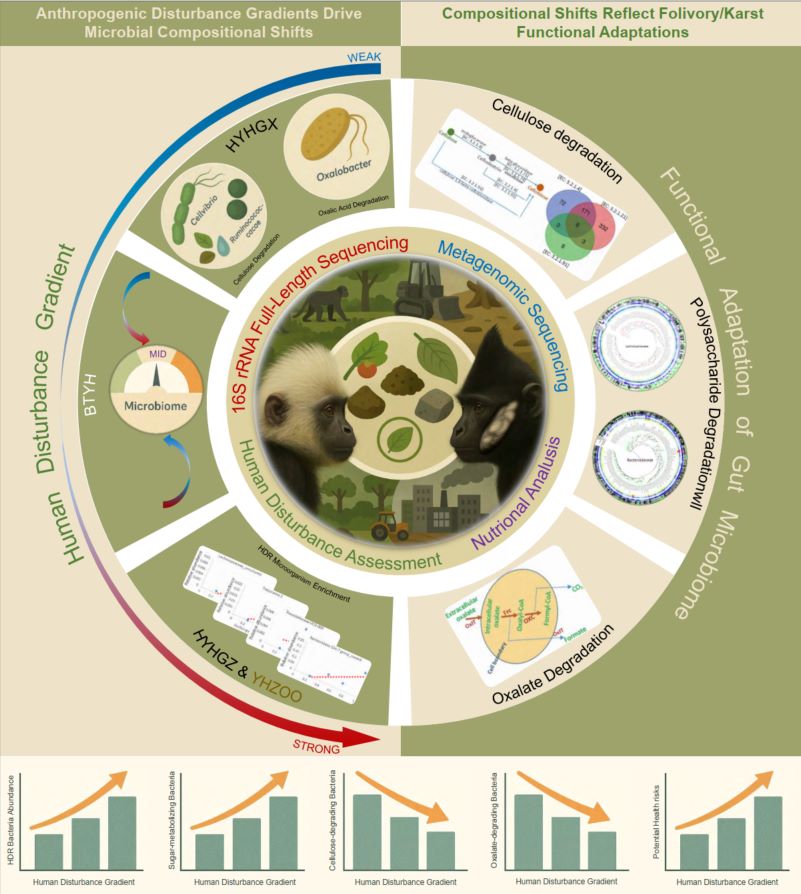
The transmission history and adaptive evolution of Mycobacterium tuberculosis complex (MTBC) in China remain underexplored despite its remarkably low diversity and enduring public health burden. Here, we analyzed 23,873 whole genome sequences of MTBC to reconstruct its spread timeline and routes, population dynamics, and host adaptation patterns in China. The Bayesian coalescent models revealed the recurrent introductions of four MTBC sub-lineages (L2.2, L4.2, L4.4, and L4.5) during 1000–400 years ago, by European-Chinese transmission networks, which might have triggered the formation of their local genetic clades in China. These local clades underwent three rapid population expansions that temporally aligned with historical climate cooling events and warfare, and displayed convergent adaptation, including shared mutations and structural variations in macrophage-resistance genes and enhanced genetic diversity in T cell epitopes. The historical mutation rate estimations and neutral mutation simulations indicated that these local clades experienced pronounced macrophage-induced pressures, likely operative over the past two centuries. The DNA information entropy analysis further revealed their adaptive evolution signatures clustered within macrophage-resistance pathways. The gene Rv0801, computationally predicted to exhibit the most prominent adaptive signatures despite its uncharacterized function, was confirmed through recombinant strain infection to enhance intracellular survival in human macrophages. This integrative approach reveals MTBC's evolutionary trajectory in China, provides novel methods for quantifying selection pressures and detecting adaptive evolution signals in prokaryotes, and constructs a comprehensive framework for investigating pathogen transmission dynamics and host adaptation mechanisms.
Soil pH modulates microbial nitrogen allocation in soil via compositional and metabolic shifts across forests in Japan
- 18 September 2025
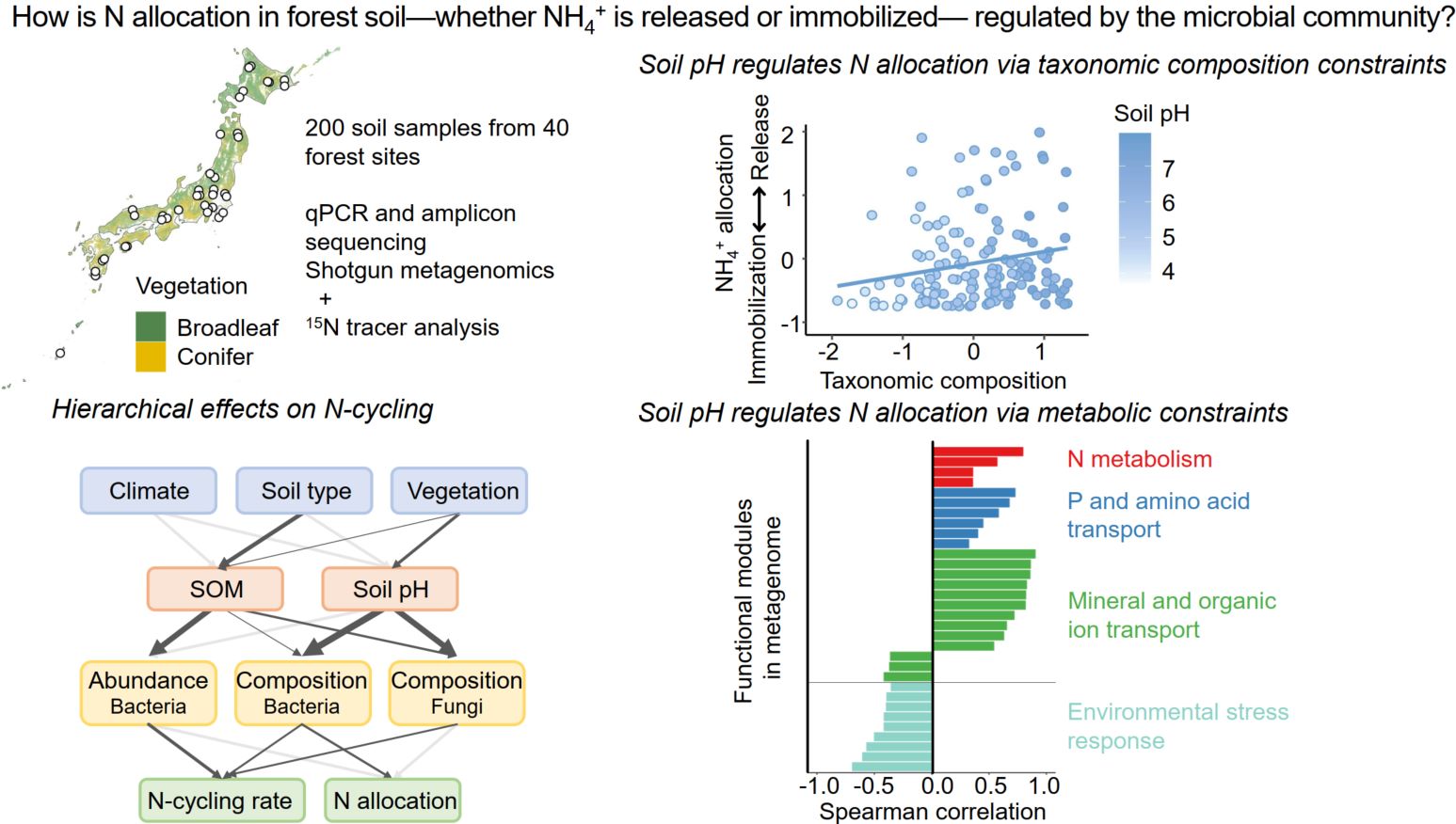
This study reveals how soil pH regulates microbial N allocation in forest soils across Japan. We show that microbial abundance controls the overall rate of N cycling, while microbial composition and metabolic potential determine whether ammonium is released or retained. In acidic soils, stress-tolerant microbes dominate, promoting N retention and suppressing nitrification. In near-neutral soils, energy-oriented taxa are enriched in metabolic pathways for ammonium release and nitrification, enhancing N mobility. These findings highlight a hierarchical structure—from pedogenic factors to soil properties to microbial community structure and function—that governs ecosystem-level N cycling, with soil pH acting as a key environmental filter.
Multiomics analyses reveal that PRMT5 regulates membrane transport and cholesterol synthesis in white adipocytes
- 05 October 2025
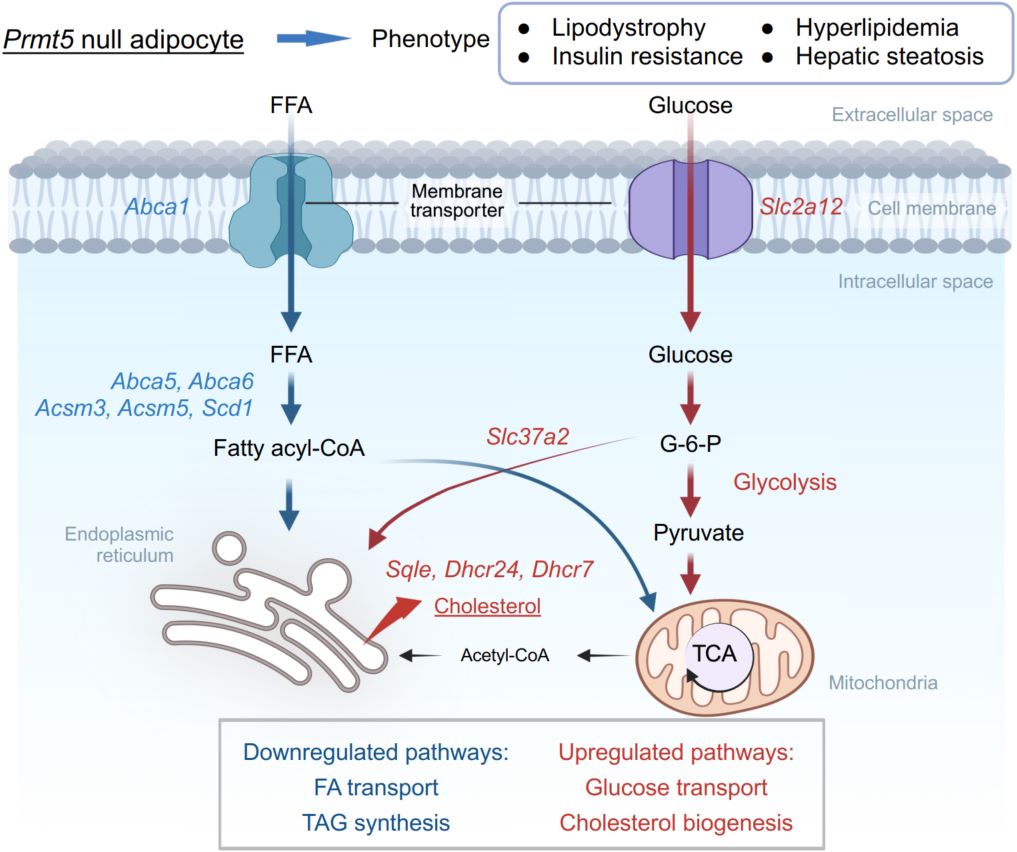
The adipose tissue (AT) is a main regulator of systemic energy homeostasis, and AT dysfunction leads to insulin resistance and other metabolic complications. Protein arginine methyltransferase 5 (PRMT5) catalyzes symmetrical dimethylation of arginine residues to modulate protein stability and function. Besides its well-studied oncogenic functions, PRMT5 has recently been shown to play a physiological role in AT through poorly understood mechanisms. Here, we explore the function of PRMT5 in AT through unbiased RNA sequencing and lipidomic analyses of AT in wild-type and adipocyte-specific Prmt5 knockout (Prmt5AKO) mice. Transcriptomic profiling revealed that Prmt5AKO alters the expression of genes related to metabolism and membrane transport. Specifically, Prmt5AKO induces genes enriched in glucose transport and glycolysis pathways, while suppressing genes encoding fatty acid (FA) transporters. Lipidomics analysis showed altered compositions of triacylglycerols (TAGs), fatty acids (FAs), and glycerophospholipids. Additionally, Prmt5AKO promotes cholesterol biogenesis, associated with hyperlipidemia and hepatic steatosis in mice. These multiomics approaches uncover previously unappreciated roles of PRMT5 as an epigenetic regulator of metabolic homeostasis via coordinating membrane transport, balancing glucose and FA metabolism, and promoting cholesterol biosynthesis. This study highlights a novel mechanism by which protein methylation regulates systemic energy balance.
Stigmasterol attenuates atherosclerosis by inhibiting inflammatory signaling and foam cell formation
- 01 October 2025
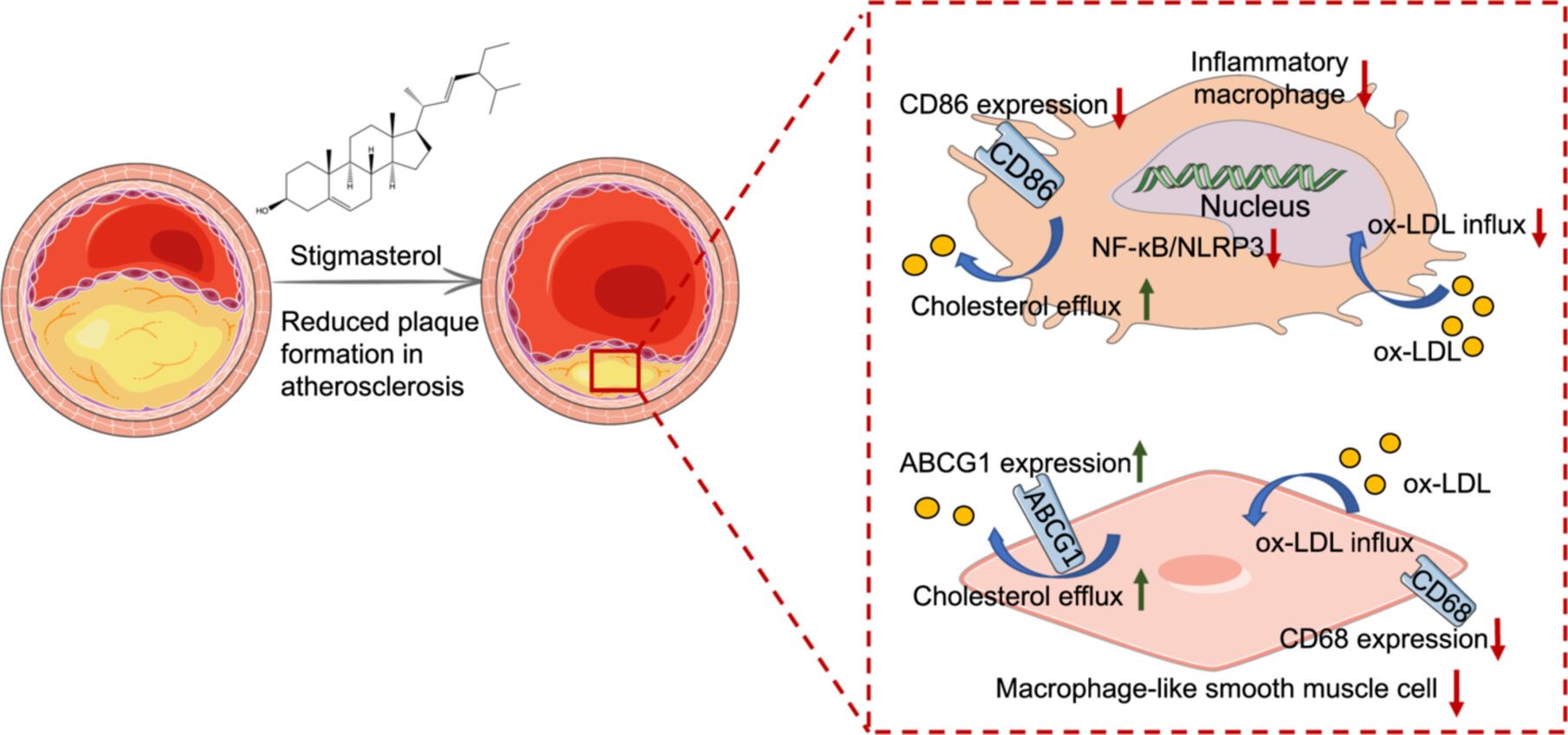
Stigmasterol mitigates atherosclerotic plaque formation by modulating foam cell phenotypic alterations and cholesterol metabolism. Stigmasterol inhibits inflammatory CD86+ macrophage polarization via the NF-κB/NLRP3 pathway and downregulates CD68 expression in smooth muscle cells (SMCs), contributing to reduced vascular inflammation. It also enhances cholesterol efflux and suppresses oxidized low-density lipoprotein (ox-LDL) uptake in foam cells. These findings offer potential therapeutic strategies for atherosclerosis prevention and treatment.
The combined influence of microbiome and soil environment contributes to the chemotype differentiation in Atractylodes lancea
- 09 October 2025
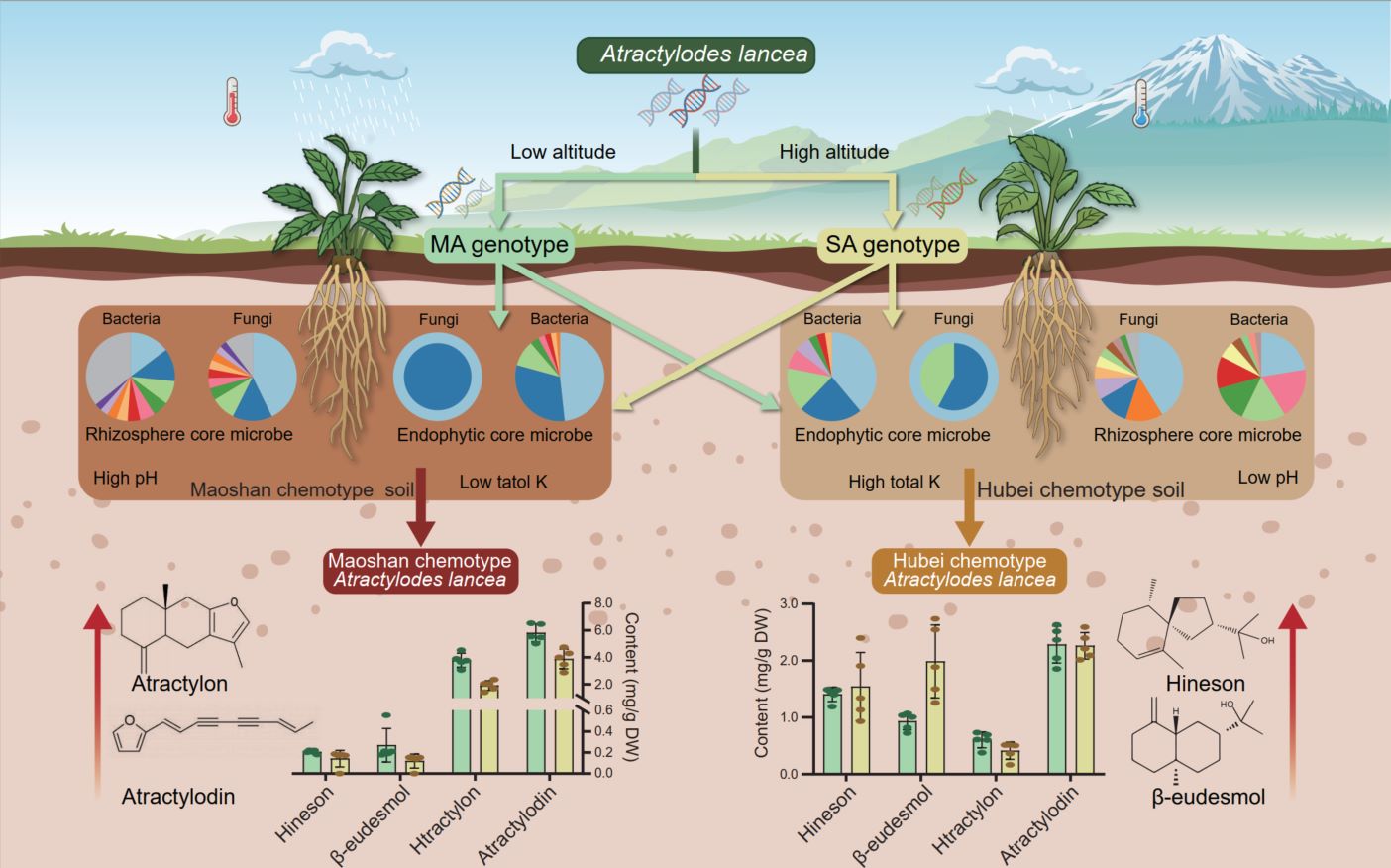
Genetic differentiation between the Maoshan-Dabie Mountains group (MA genotype) and Qinling-Taihang Mountains group (SA genotype) of Atractylodes lancea occurs along altitude and climatic gradients. High altitudes and their corresponding climatic conditions favor the formation of SA, while low altitudes and their corresponding climatic conditions favor the formation of MA. The chemotype differentiation of the geo-authentic Maoshan chemotype (MSA, rich in atractylon and atractylodin) and the non-authentic Hubei chemotype (HBA, dominated by hinesol and β-eudesmol) in A. lancea is influenced by soil chemistry and microbiota, rather than being determined by its genotype. Rhizosphere and endophytic core microbes are the driving forces behind the chemotype differentiation of A. lancea.
Electroacupuncture reshapes the microbial co-occurrence networks related to the behavioral and psychological symptoms of dementia in Alzheimer's disease
- 02 July 2025
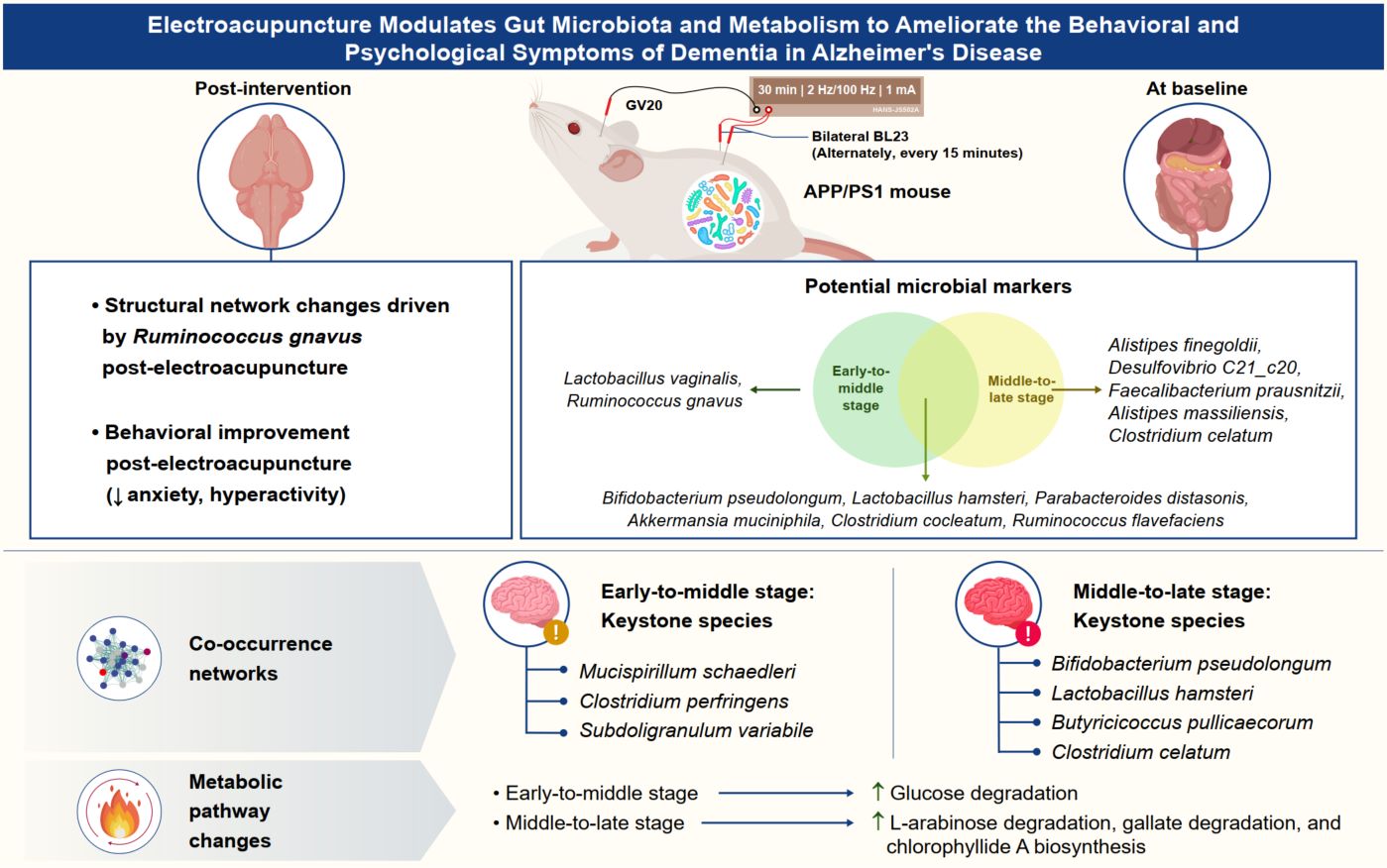
Microbial keystone species and gut microbiota composition are highly variable during the pathological development of the behavioral and psychological symptoms of dementia (BPSD) in Alzheimer's disease (AD). Age stratification reveals stage-specific gut microbial signatures in AD-related BPSD. This study highlights the efficacy of electroacupuncture in regard to altering the intestinal microbial landscape in AD-related BPSD and provides novel insights into the application of phased targeted electroacupuncture interventions in the future.
BioMGCore: A toolkit for detection of biological metabolites in microbiome
- 22 June 2025
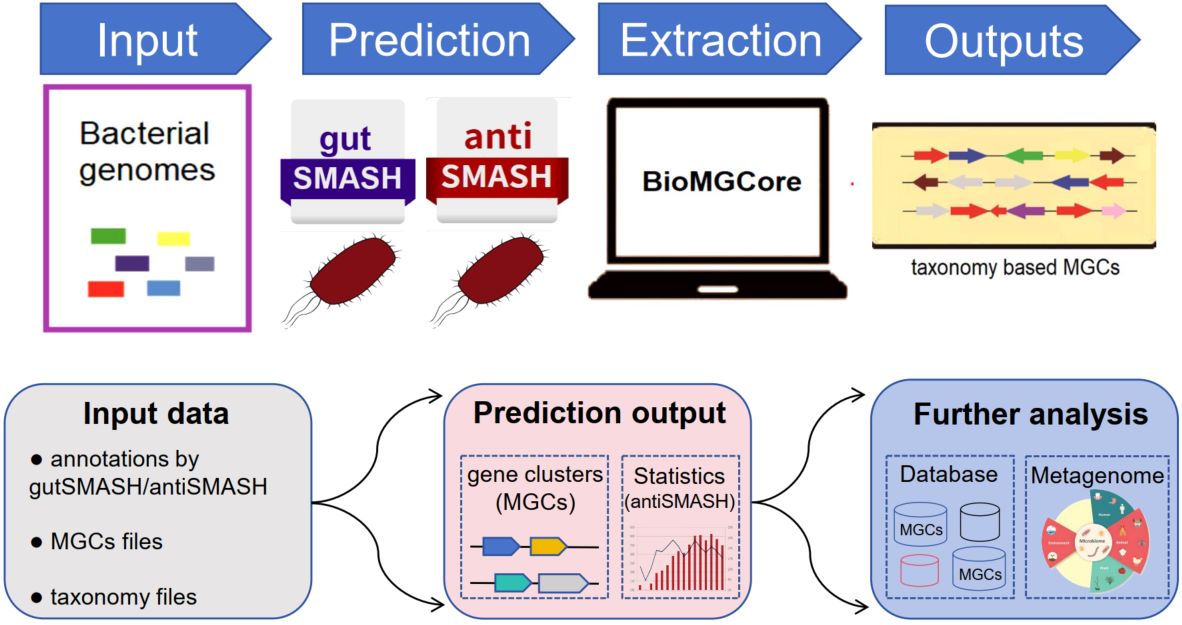
The keyword matching and gene proximity principles were used to accurately identify core gene clusters in microbial genomes. The metabolic gene clusters can be classified into different taxonomic groups according to Domain, Phylum, Class, Order, Family, Genus, and Species. BioMGCore can achieve batch statistics for secondary metabolites prediction.
TBCancer: A database for exploring characteristics and functions of tissue-biased genes in cancer
- 29 June 2025
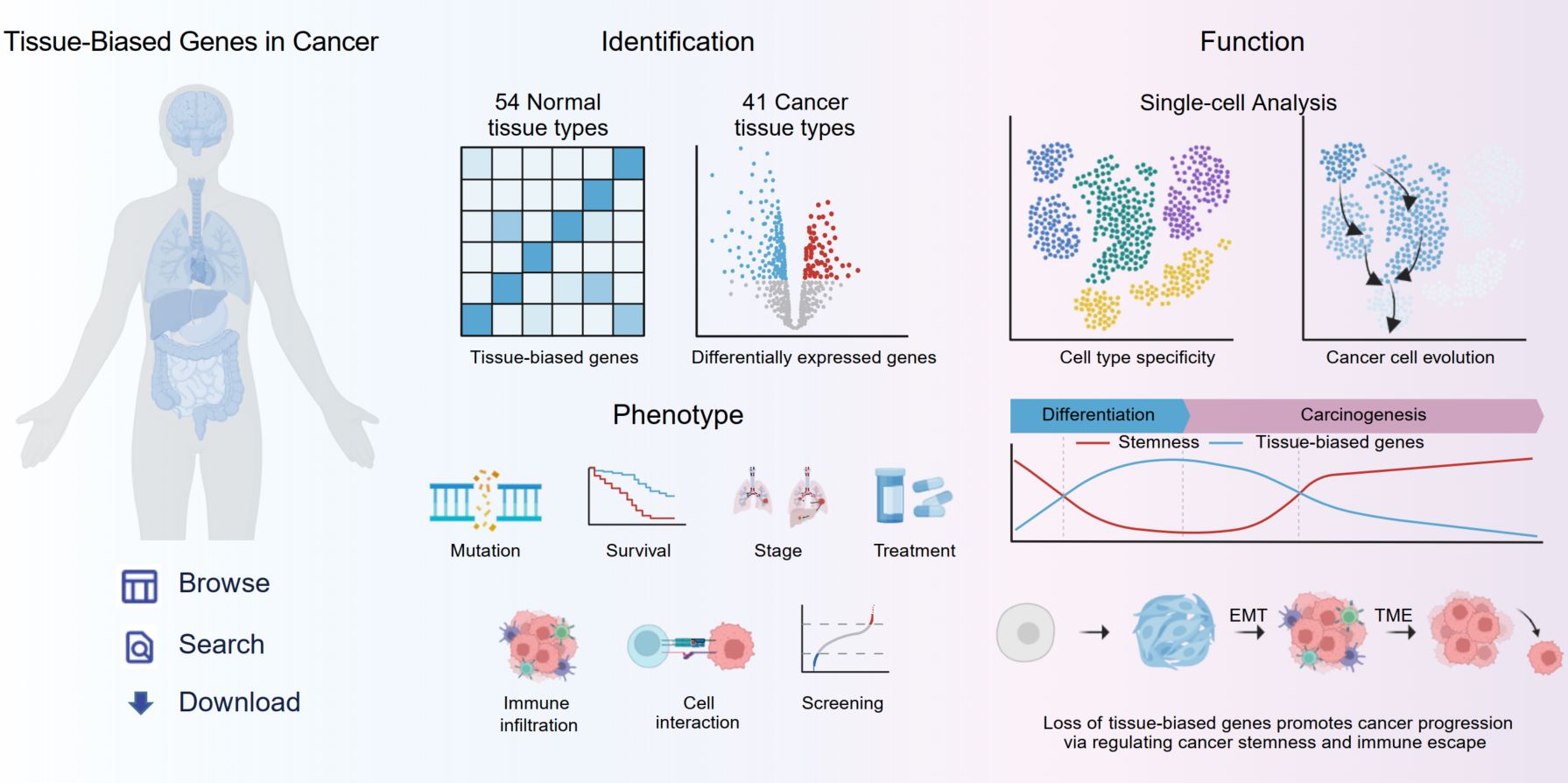
This study defined 10,921 tissue-biased genes across 54 normal tissues and 41 cancer types. Tumor-associated tissue-biased genes exhibit downregulation, mutations, and epigenetic modifications, correlating with poor clinical outcomes. Their inactivation promotes tumorigenesis by enhancing stemness and immune evasion, highlighting their value as prognostic biomarkers and therapeutic targets. To facilitate research, we developed a database integrating multi-omics data on these genes for mechanistic and therapeutic exploration.
Saliva MicroAge: A salivary microbiome based machine learning model for noninvasive aging assessment and health state prediction
- 08 July 2025
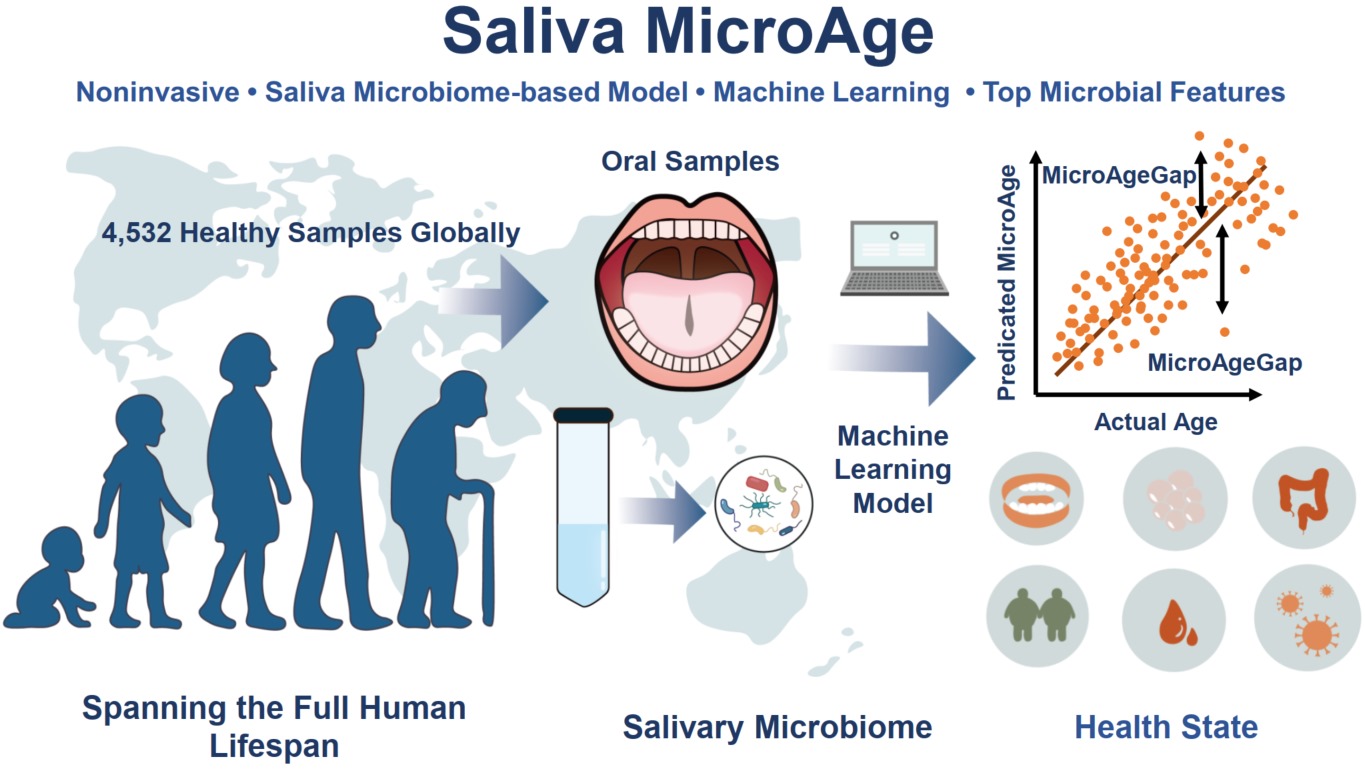
Saliva MicroAge is a machine learning-based model designed to estimate biological age and assess health status using globally sourced salivary microbiome data. Trained on 4532 healthy samples, the model achieves high accuracy in predicting chronological age and captures health-related deviations (MicroAgeGap) in various diseases. Taxonomic and functional analyses of key microbial features reveal biological relevance to aging processes, offering a noninvasive and scalable approach for aging monitoring and precision health assessment.
HiFi based metagenomic assembly strategy provides accuracy near isolated genome resolution in MAG assembly
- 08 July 2025
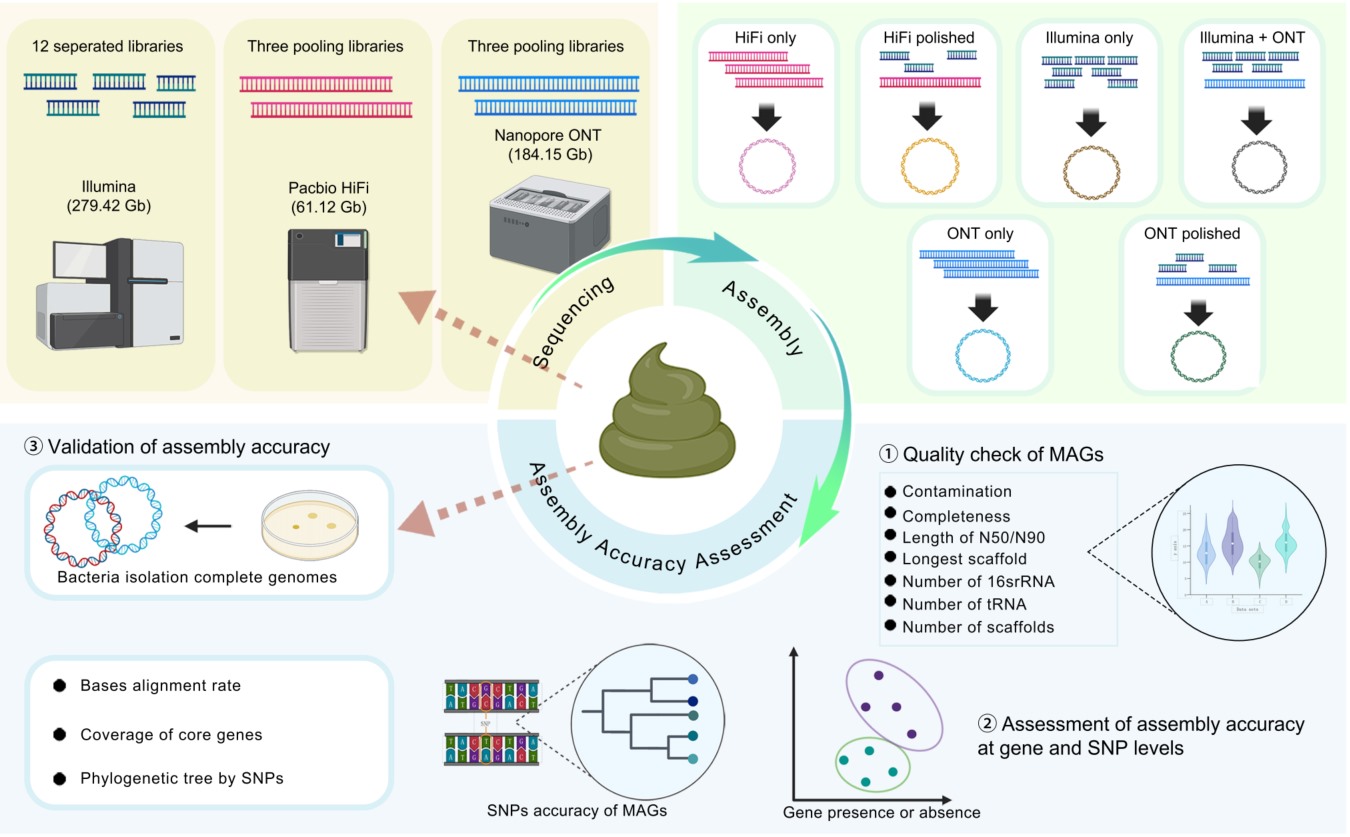
Recovering high-contiguity, circular bacterial genomes from complex microbiomes (e.g., gut) is challenged by limitations of short-read and error-prone long-read sequencing. This study comprehensively compares PacBio High-Fidelity (HiFi) sequencing-based metagenome-assembled genomes (MAGs) against Illumina MAGs, Oxford Nanopore Technologies (ONT) MAGs, and isolate whole-genome sequencing genomes from the same sample. HiFi sequencing yielded 31 high-quality MAGs, including 10 complete circular genomes. HiFi MAGs demonstrated significantly higher completeness, continuity, and lower contamination than Illumina or ONT MAGs (p-adj < 0.05).Crucially, HiFi MAGs exhibited closer genomic proximity to corresponding isolates at both single-nucleotide polymorphism and gene presence/absence levels. This benchmarking establishes HiFi as a robust approach for generating MAGs rivaling isolated genome quality, providing critical insights for accurate microbial genomic studies.
Distinctive metabolic disturbances associated with redox homeostasis, nervous and hormonal functions during gut microbial enrichment upon polystyrene microplastic exposure
- 23 July 2025
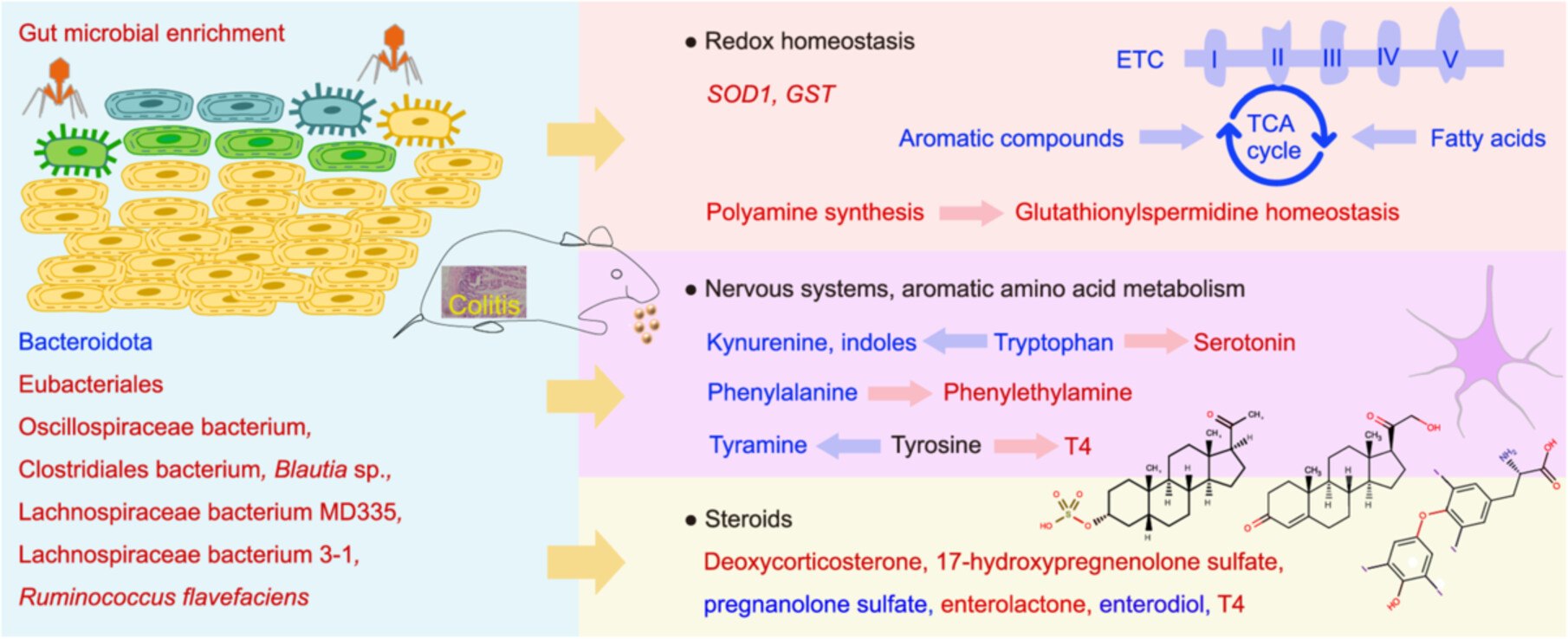
Microplastic-induced gut microbial enrichment was dominated by bacteria within Eubacteriales, correlated with the virome, and accompanied by colitis. The polyamine synthetic pathway was activated to maintain glutathionylspermidine homeostasis, concurrent with decreases in pathways involved in the production of energy and reactive oxygen species under microplastic exposure. Tryptophan-serotonin, phenylalanine-phenylethylamine, and tyrosine-thyroxine pathways increased, whereas tryptophan-kynurenine, tryptophan-indole, and tyrosine-tyramine pathways decreased under microplastic exposure. Enterolactone synthesis and cholesterol-derived hormone synthesis were increased under microplastic exposure. Bacteria within Eubacteriales (e.g., Oscillospiraceae bacterium and Clostridiales bacterium) contributed most to metabolic disturbances under microplastic exposure.
RTTAP: Empowering metatranscriptomic data analysis with a read-based total-infectome taxonomic solution
- 29 July 2025
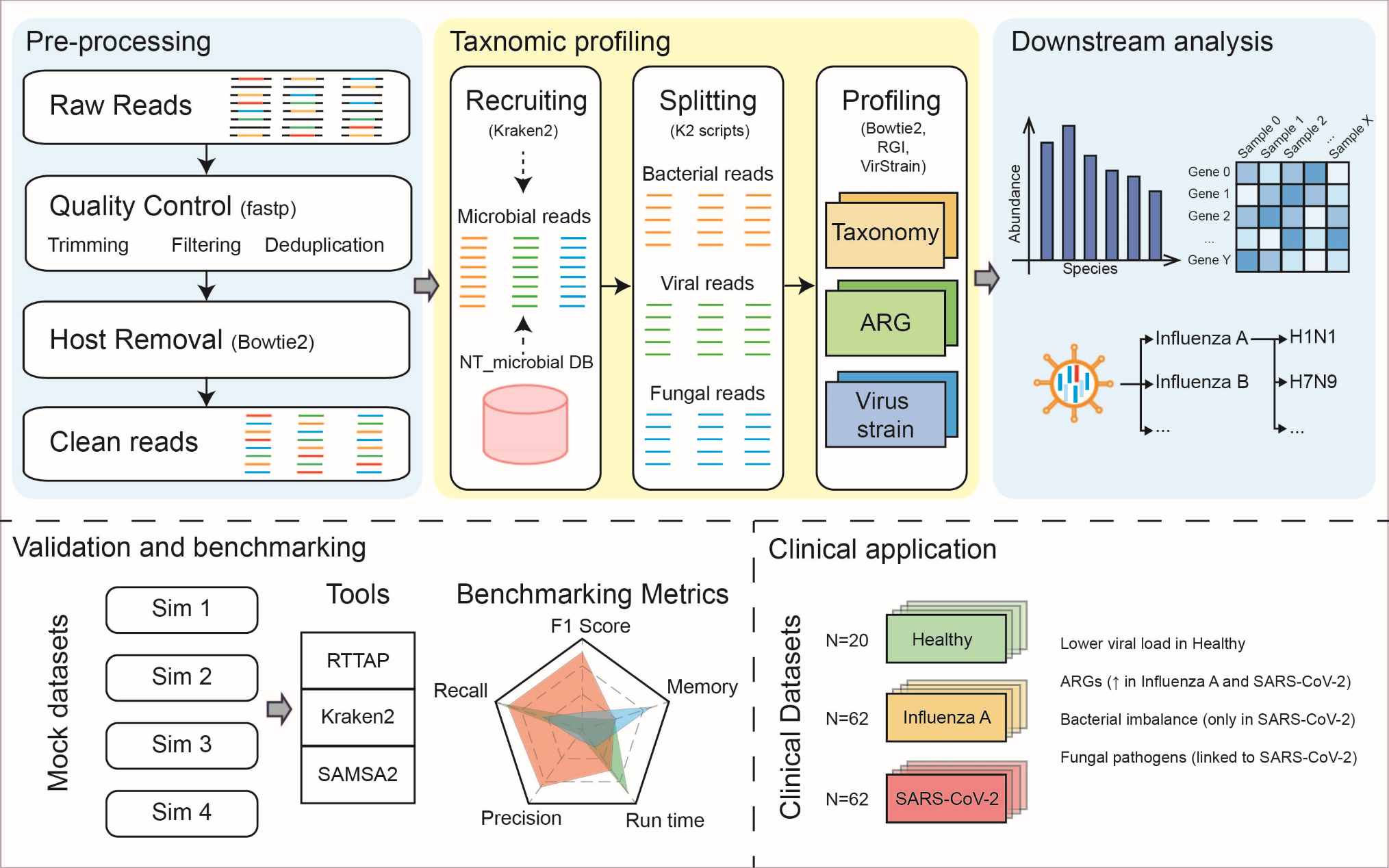
Metatranscriptomic data analysis is a complex task due to its sheer volume and the need for sophisticated bioinformatics tools. To address this, we developed the Read-based Total-infectome Taxonomic Analysis Pipeline (RTTAP), an automated pipeline for metatranscriptomic data analysis that eliminates the need for users to manually select databases, tools, or parameters. RTTAP provides a comprehensive solution for “total-infectome” analysis, enabling simultaneous detection of viruses, bacteria, and fungi. Additionally, RTTAP delivers detailed functional profiling of antibiotic resistance genes (ARGs) and high-resolution viral strain analysis, offering researchers a powerful tool for advanced metatranscriptomic studies. The pipeline's performance was validated using both simulated and real clinical metatranscriptomic datasets, demonstrating high accuracy in taxonomic classification and relative abundance estimation.
GISDD: A comprehensive global integrated sequence and genotyping database platform for dengue virus, facilitating a stratified coordinated surveillance strategy
- 27 August 2025
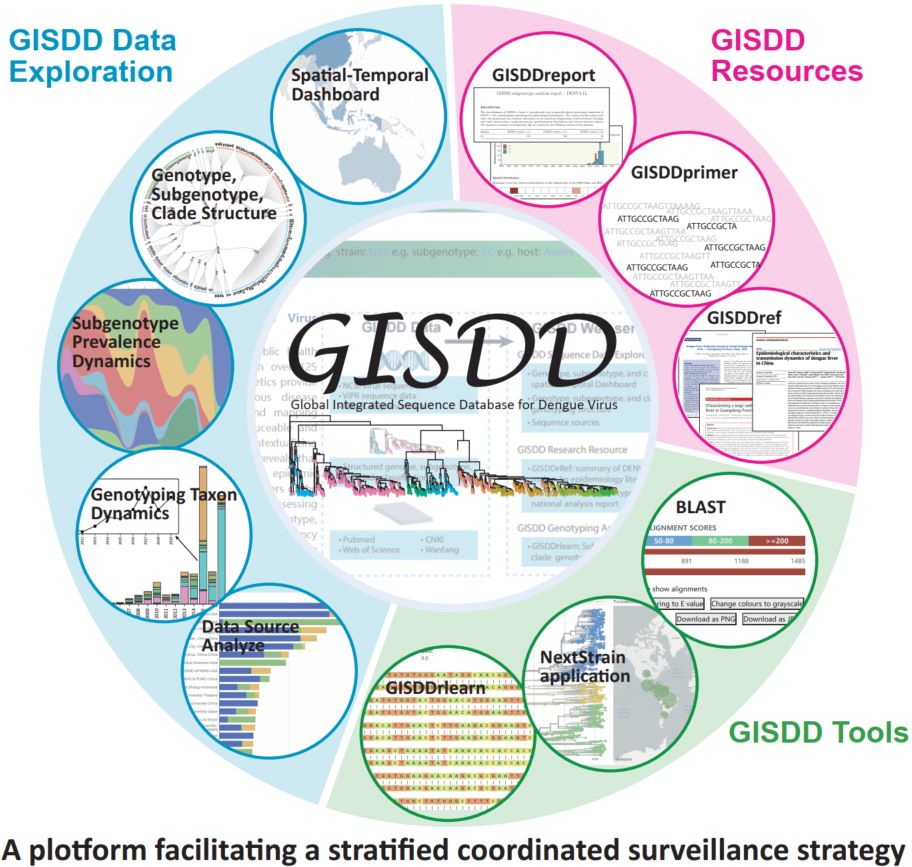
GISDD, the Global Integrated Sequence and Genotyping Database for Dengue Virus (DENV), provides an integrated online analysis platform encompassing tools such as GISDDrlearn, GISDDprimer, and GISDDref, enabling swift identification and tracing of well-established DENV genotypes, subgenotypes, and clades, thereby facilitating a stratified global coordinated surveillance strategy.