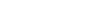• Your research will be open access, resulting in more downloads, shares and cites.
• There’s no cost to you. Article publication fees are currently waived.
• Video submissions make it easier for readers to access and digest content.
• Open data and codes will help further advance our knowledge and research and drive impact in the field.
• Figures are polished for the highest quality publication.
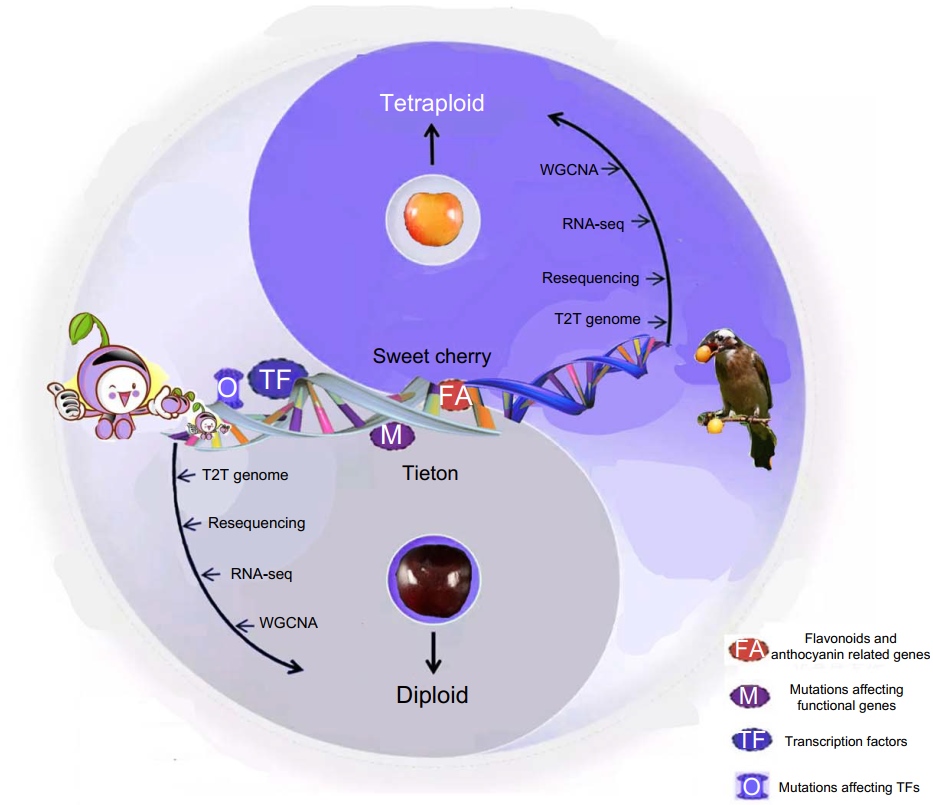
Oleoylethanolamide regulates intestinal stem cell activity and villus size via PPARα signaling pathway
- 26 January 2026
First use of a natural swine model with lipid metabolism to directly link lipid differences to a quantifiable intestinal villus height phenotype. Identified the phospholipid-derived oleoylethanolamide, rather than canonical fatty acids, as the endogenous primary ligand that activates peroxisome proliferator-activated receptors α (PPARα) to enlarge villi. Established and validated a PPARα-oxidative phosphorylation–stem cell energy switch axis, offering a druggable metabolic node that couples nutrient status to morphogenesis.
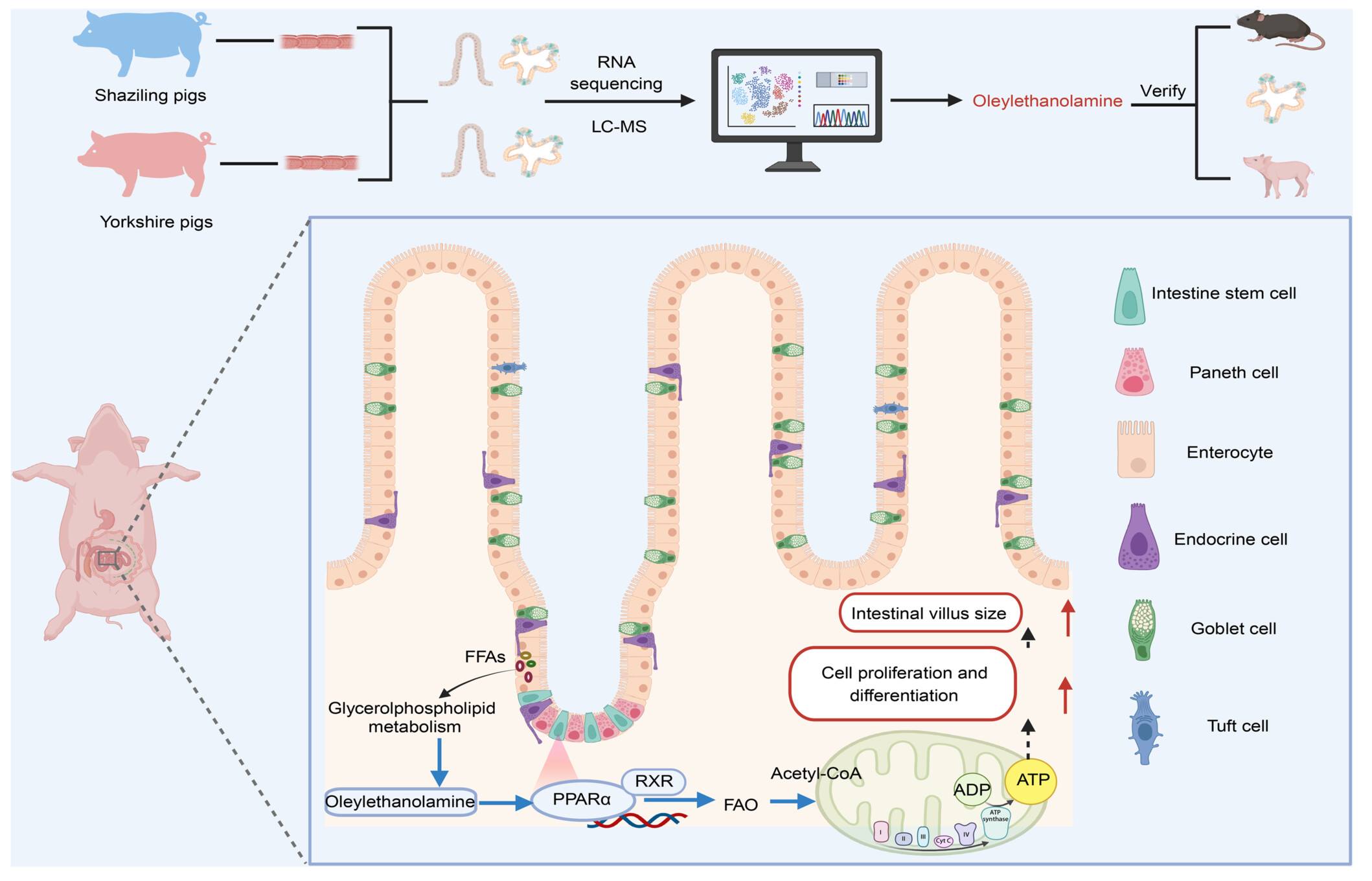
ECCFP: A consecutive full pass-based bioinformatic analysis for eccDNA identification from long-read sequencing data
- 03 February 2026
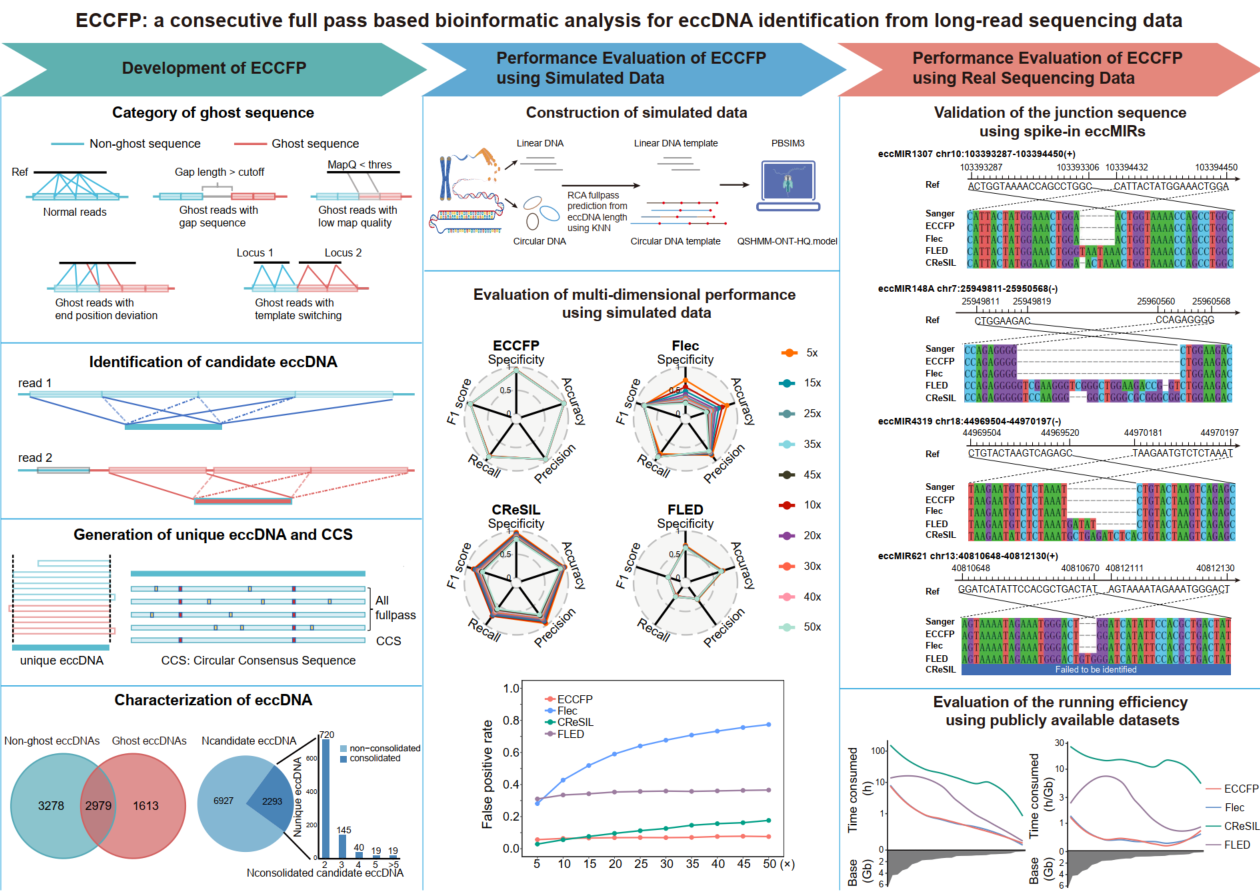
A new bioinformatics pipeline called ECCFP has been developed to improve the detection of extrachromosomal circular DNA (eccDNA) from long-read sequencing data. ECCFP uses all consecutive full passes from individual reads for candidate eccDNA identification and consolidates candidate eccDNAs to generate accurate unique eccDNA. To thoroughly evaluate the performance of ECCFP, 6 simulated datasets, 2 real sequencing datasets with spiked-in artificial synthesized eccDNAs and 29 publicly available real sequencing datasets, were applied. The results demonstrate that ECCFP markedly reduces the false positive rate, significantly increases the number of unique eccDNAs detection, achieves higher accuracy in identifying junction positions and circular consensus sequences, and also shortens the runtime, when compared to other three eccDNA analysis pipelines. Overall, ECCFP provides a more efficient and accurate bioinformatic analysis for eccDNA detection from long-read sequencing data, which might push the research forward on eccDNA as a biomarker in clinical settings.
The evolutionary landscape and serotypic dynamics of avian infectious bronchitis virus from spike protein
- 19 January 2026
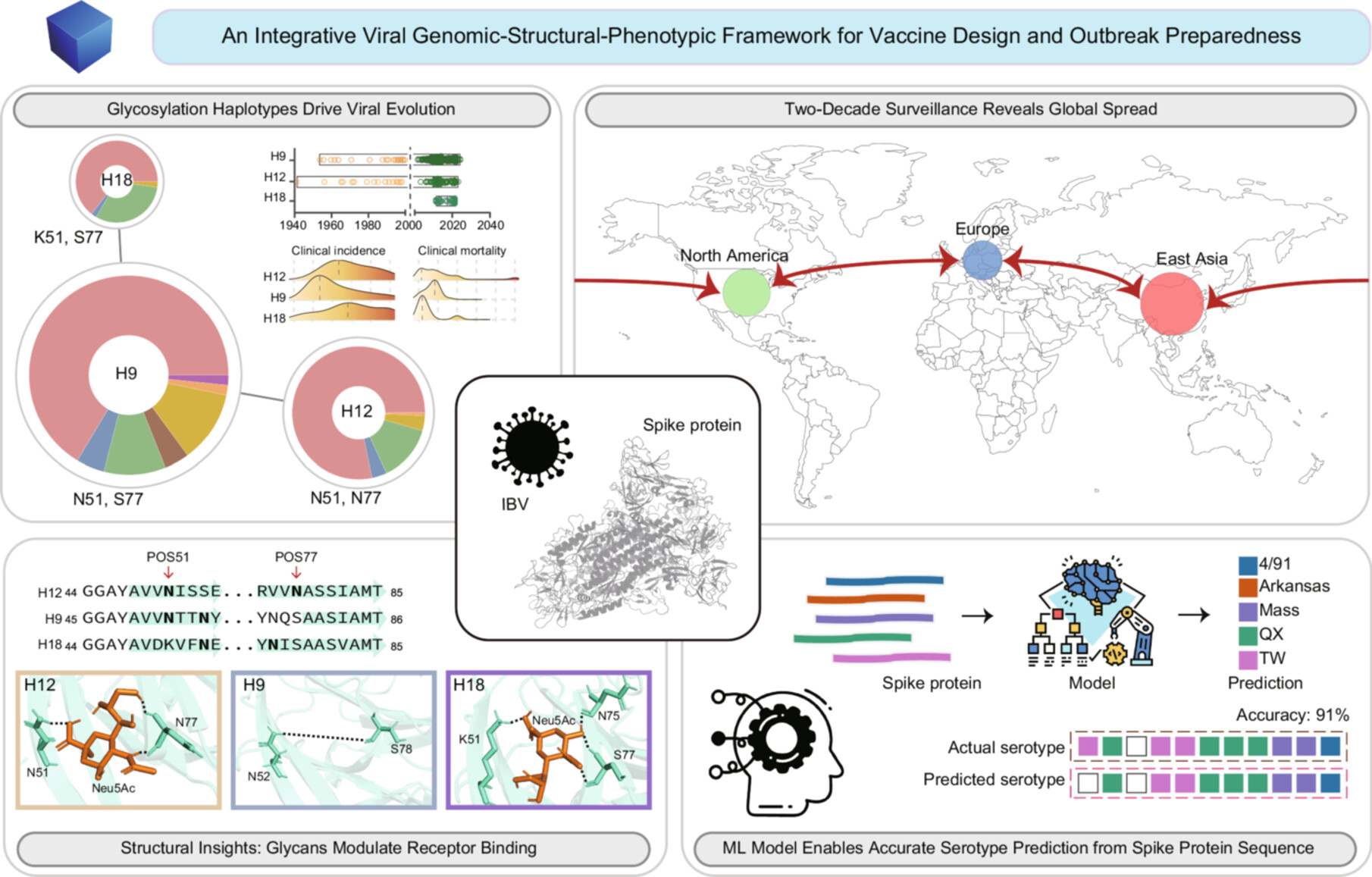
This study integrates two decades of surveillance with genomic and structural analyses to decipher how spike protein glycosylation haplotypes drive avian coronavirus evolution. We uncover how specific glycosylation patterns associate with receptor-binding affinity, shape global transmission dynamics, and correlate with clinical outcomes. Furthermore, we develop a machine learning framework for highly reliable serotype prediction directly from spike protein sequences. This multidisciplinary work establishes a scalable “viral genomic-structural-phenotypic” framework to forecast coronavirus evolution and guide vaccine design.
Stratification of chronic rhinosinusitis with nasal polyps by distinct sinonasal microbial communities and their clinical and pathological correlations
- 06 January 2026
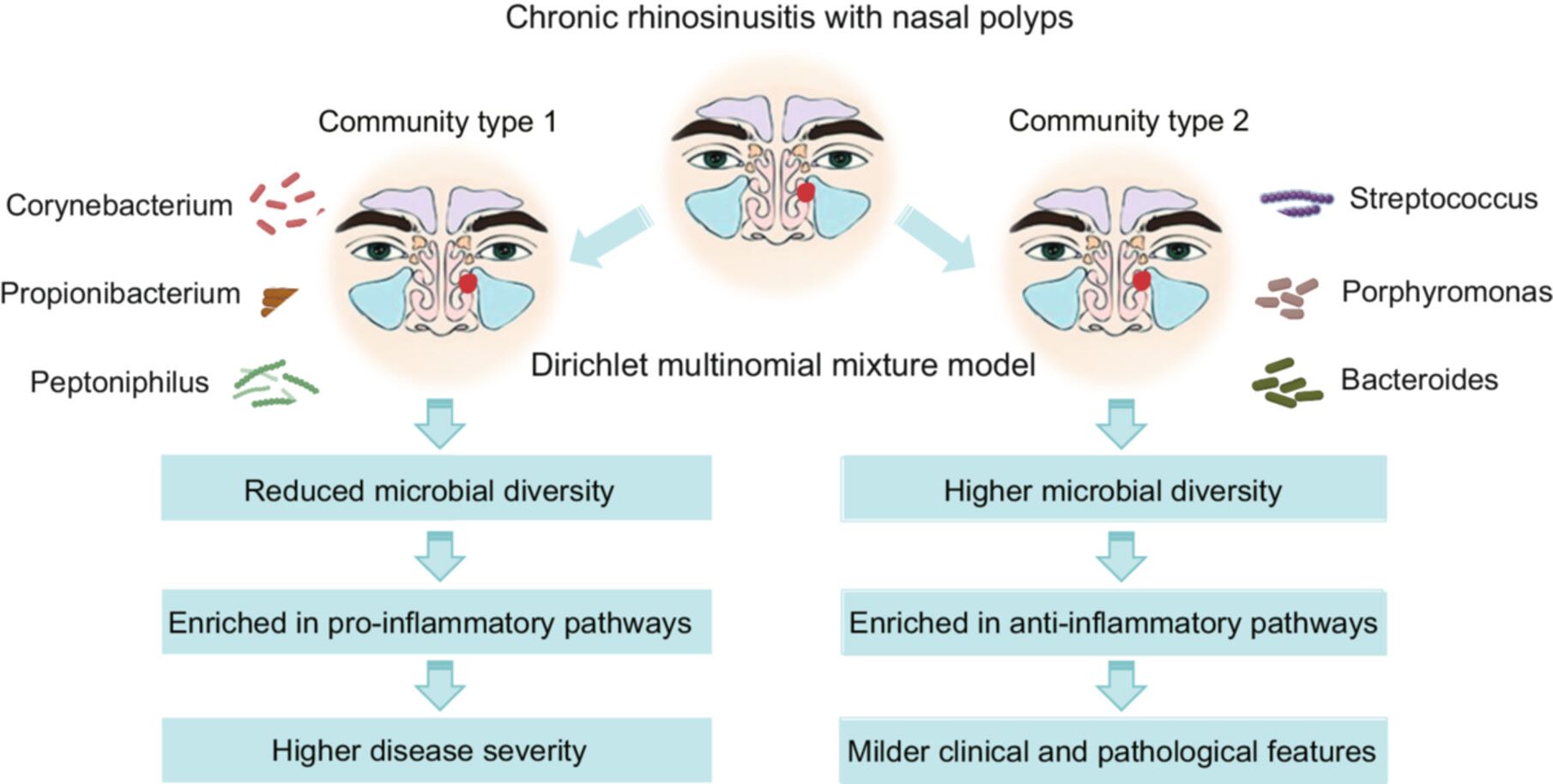
Chronic rhinosinusitis with nasal polyps (CRSwNP) is a heterogeneous inflammatory condition. Dysbiosis is commonly observed. This study identified distinct sinonasal microbial communities in CRSwNP patients and explored their clinical and pathological associations. Two distinct microbial communities were identified: community type 1 (CT1), dominated by Corynebacterium, Propionibacterium, and Peptoniphilus; and community type 2 (CT2), dominated by Streptococcus, Porphyromonas, and Bacteroides. CT1 exhibited significantly reduced microbial diversity and was enriched in pro-inflammatory pathways, whereas CT2 showed higher microbial diversity, with pathways supporting anti-inflammatory. Clinically, CT1 patients demonstrated higher disease severity and increased eosinophil infiltration, compared with CT2. CRSwNP patients can be stratified into two distinct microbial communities, each with specific clinical and pathological profiles. Microbial-based stratification may inform tailored therapeutic strategies to address the diverse clinical and pathological manifestations of CRSwNP.
Diversity, transfer potential, and transcriptional activity of virus-carried antibiotic resistance genes in global estuaries
- 07 January 2026
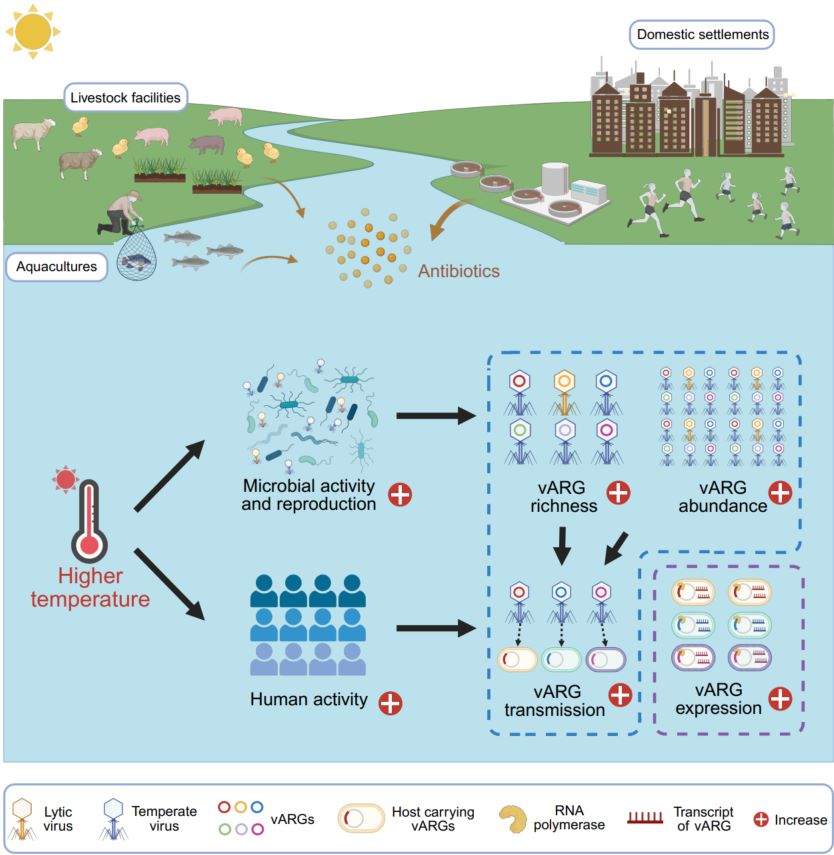
Estuaries are vital hotspots for antibiotic resistance genes (ARGs) due to substantial antibiotic pollution. Although viruses have been proposed as key reservoirs and important disseminators of ARGs in environments, their contribution to the estuarine antibiotic resistome remains largely unknown. Here, by constructing a Global Estuarine Virome dataset from 534 worldwide estuarine metagenomes, we found that estuarine viruses, particularly temperate viruses, carried abundant and diverse ARGs, and over half of the virus-carried ARGs (vARGs) were actively expressed in estuarine environments. Further analysis based on sequence similarity and minimal evolutionary distance revealed that vARG-carrying viruses targeted a wide range of prokaryotes and likely facilitated horizontal gene transfer of vARGs between viruses and prokaryotes. Importantly, elevated temperatures and coastal human activities could significantly increase the richness, abundance, transmission risk, and relative expression of estuarine vARGs. These findings provide important implications for managing the estuarine antibiotic resistome.
Resistome flow in global landfill systems
- 06 January 2026

Antibiotic resistance genes (ARGs) represent a critical global public health threat. Effective resistome management within the One Health framework requires a comprehensive understanding of the sources and movement of ARGs across environmental media. This study provides a detailed analysis of resistome characteristics and dynamics in global landfill systems. Landfill systems were found to harbor diverse and highly mobile ARGs, with multidrug-resistant genes (MDRGs) comprising 32.63% of total ARGs, and comparable or even higher levels of associated ARG subtypes than in other environmental media, such as acidic mine wastewater. The ARG density on plasmids was 3.5 times higher than that on chromosomal sequences, with five times more abundant ARG-carrying plasmid-associated contigs in landfill systems than that in wastewater. At the metagenome-assembled genome level, members of the phylum Pseudomonadota were identified as the most abundant hosts (29.72%), harboring 47.74% and 56.47% of the total ARGs and MDRGs, respectively. Genomic analysis of 35 Pseudomonas spp. strains revealed unique Pseudomonas species in landfill systems, including P. aeruginosa. Resistome flow pathways were further mapped, including inflows into landfill (via soil, wastewater, freshwater, and the human and pig gut), movement within landfills (across landfill refuse, leachate, and airborne particles), and outflows from landfills (via landfill leachate effluent, gas emissions, and refuse evacuation). The results emphasized diverse resistome development and movement within landfill systems, ultimately contributing to the environmental spread of ARGs. Thus, our study highlights the urgent need for comprehensive policies and management strategies to mitigate resistome proliferation at every stage of municipal solid waste management.
Microbial influences on immune modulation and colorectal cancer progression through combined transcriptomic and microbiomic analysis
- 04 January 2026
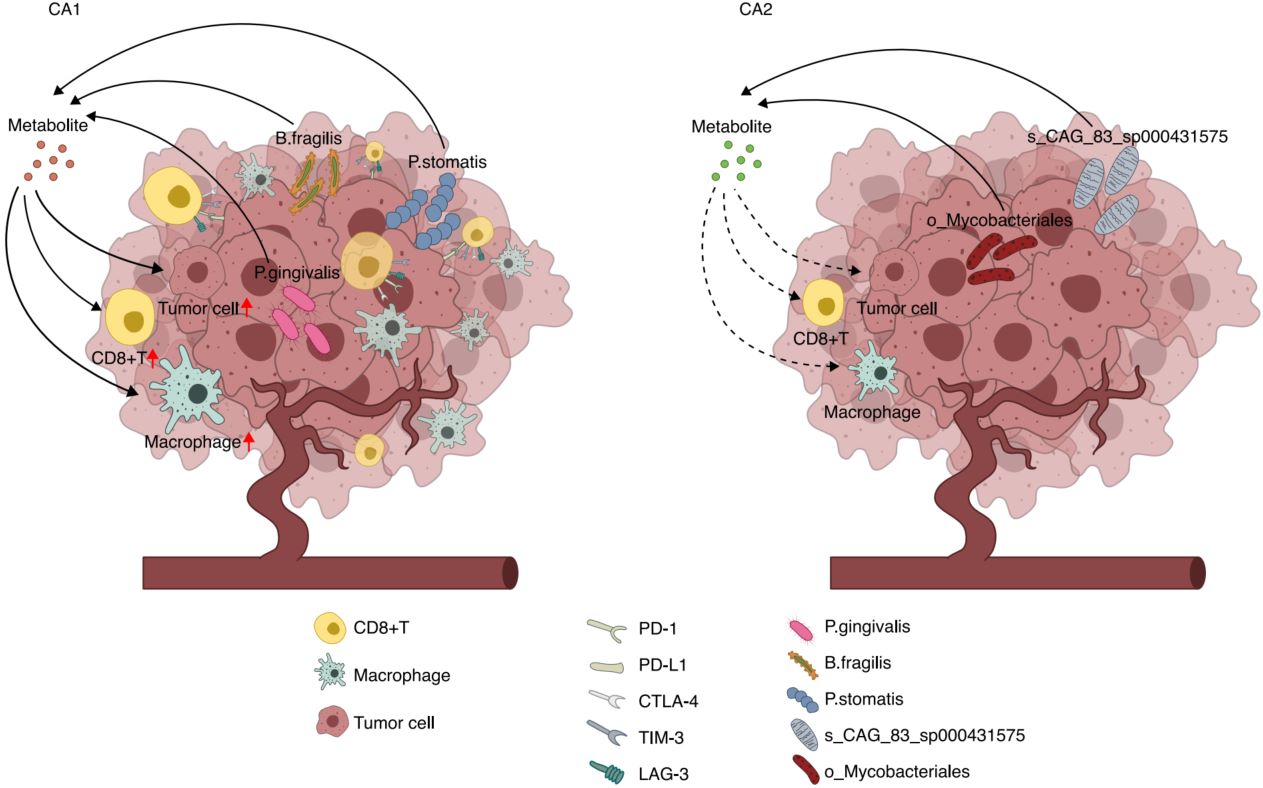
Schematic representation of the distinct tumor microenvironment characteristics in CA1 and CA2 subtypes. CA1 illustrates an immune-active tumor microenvironment characterized by elevated infiltration of immune cells, particularly CD8+T cells and macrophages, and enhanced IFN-γ signaling. This subtype shows enrichment of specific bacterial species (including P. gingivalis and B. fragilis) that contribute to immunomodulation through metabolic pathways such as γ-aminobutyric acid (GABA) and butanoate metabolism. Despite high immune activity, CA1 exhibits a poor prognosis, potentially linked to immune exhaustion, manifested by high expression of PD-1, CTLA-4, LAG-3, and TIM-3. CA2 depicts an immune-suppressed microenvironment featuring reduced immune cell infiltration, lower inflammatory signatures, and a distinct microbial composition dominated by health-associated bacteria. This tumor phenotype shows better overall survival even with reduced immune activity. Arrows indicate key molecular interactions, with solid lines representing direct effects and dashed lines indicating indirect modulation.
Ribosomal RNA operon copy number: A trait-informed framework to close the microbial cultivation gap
- 25 December 2025
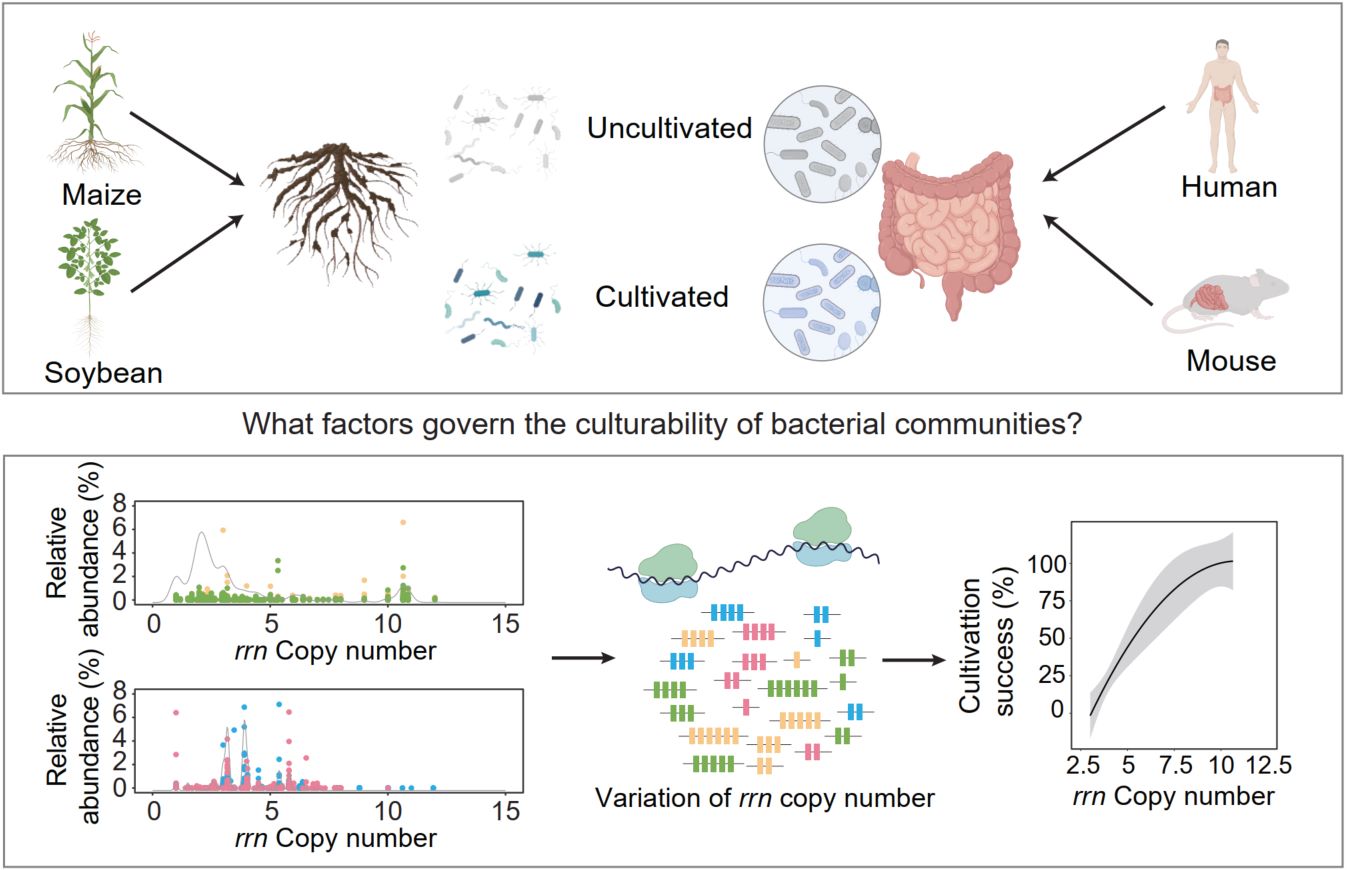
The vast majority of microbes remain uncultured, constraining functional validation and limiting microbiome-based applications. We show that ribosomal RNA operon (rrn) copy number—a stable, phylogenetically conserved genomic trait—is strongly correlated with cultivation outcomes across ecosystems. Comparative analyses of rhizosphere, human gut, and mouse gut microbiomes reveal that rhizosphere communities contain a higher proportion of high-rrn taxa, which are preferentially recovered under nutrient-rich, short-term cultivation, whereas low-rrn taxa are systematically underrepresented. By linking rrn distributions to cultivation success, we propose trait-informed strategies for media design, incubation regimes, co-culture approaches, and preservation methods. This framework provides a predictive, generalizable path to bridge sequencing and cultivation, facilitating biodiversity conservation, sustainable agriculture, and microbiome-based therapeutics.
The gut microbiome promotes the growth performance of black soldier fly larvae by detoxifying uric acid
- 24 December 2025
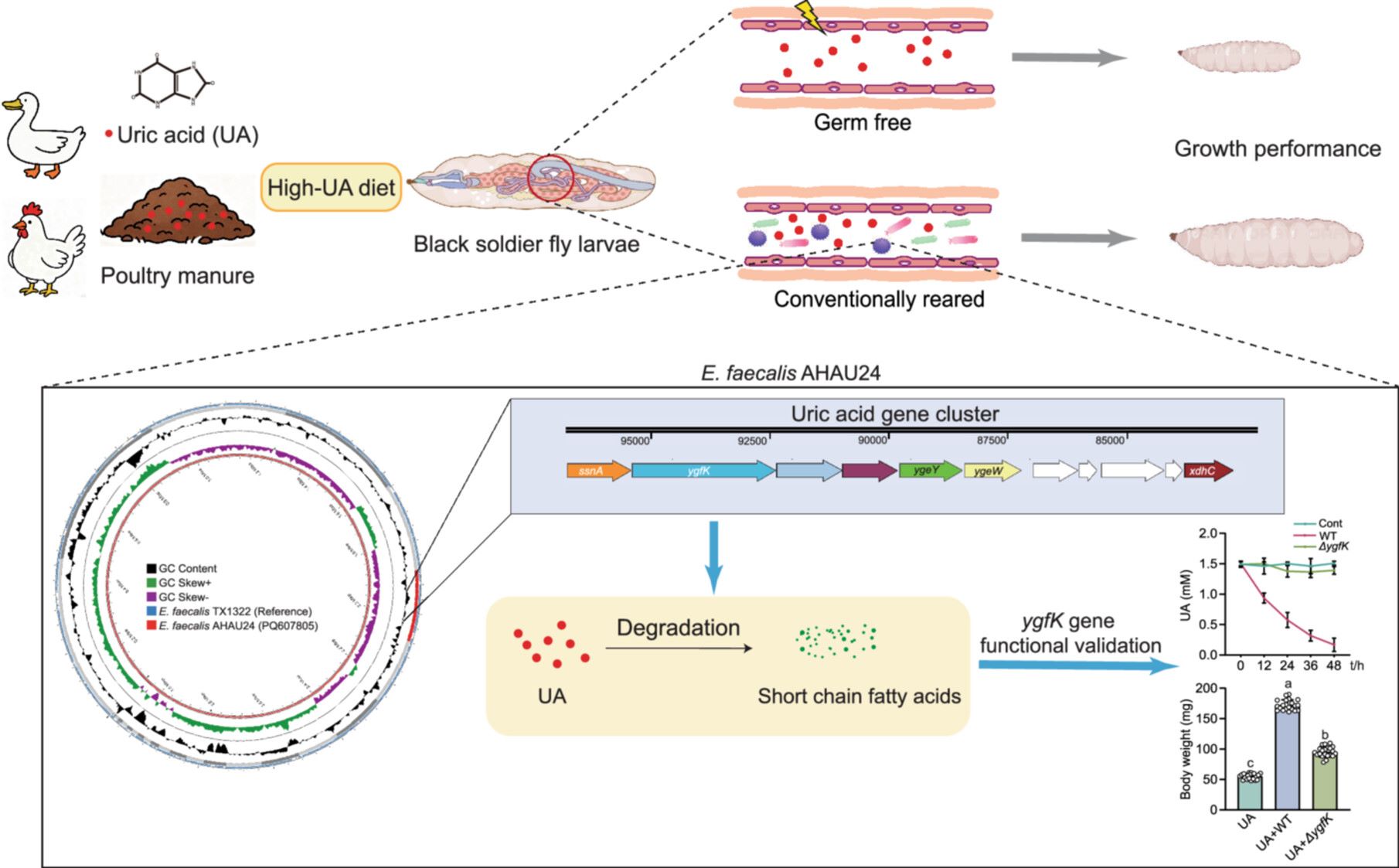
This study demonstrates the detrimental effects of exogenous uric acid (UA) on the growth of black soldier fly (BSF) larvae, highlighting the role of gut microbiota in UA degradation. We isolated UA-degrading bacterial strains associated with BSF, including Enterococcus faecalis AHAU24. Genomic analysis identified a UA degradation gene cluster in this strain, providing insights into its potential for UA catabolism. Furthermore, mono-association of germ-free larvae with E. faecalis AHAU24 confirmed that this strain alleviates UA-induced growth retardation and enhances bioconversion efficiency by degrading dietary UA.
MicrobTiSDA: A flexible R package for inferring interspecies interactions and abundance dynamics in microbiome time-series data
- 27 November 2025
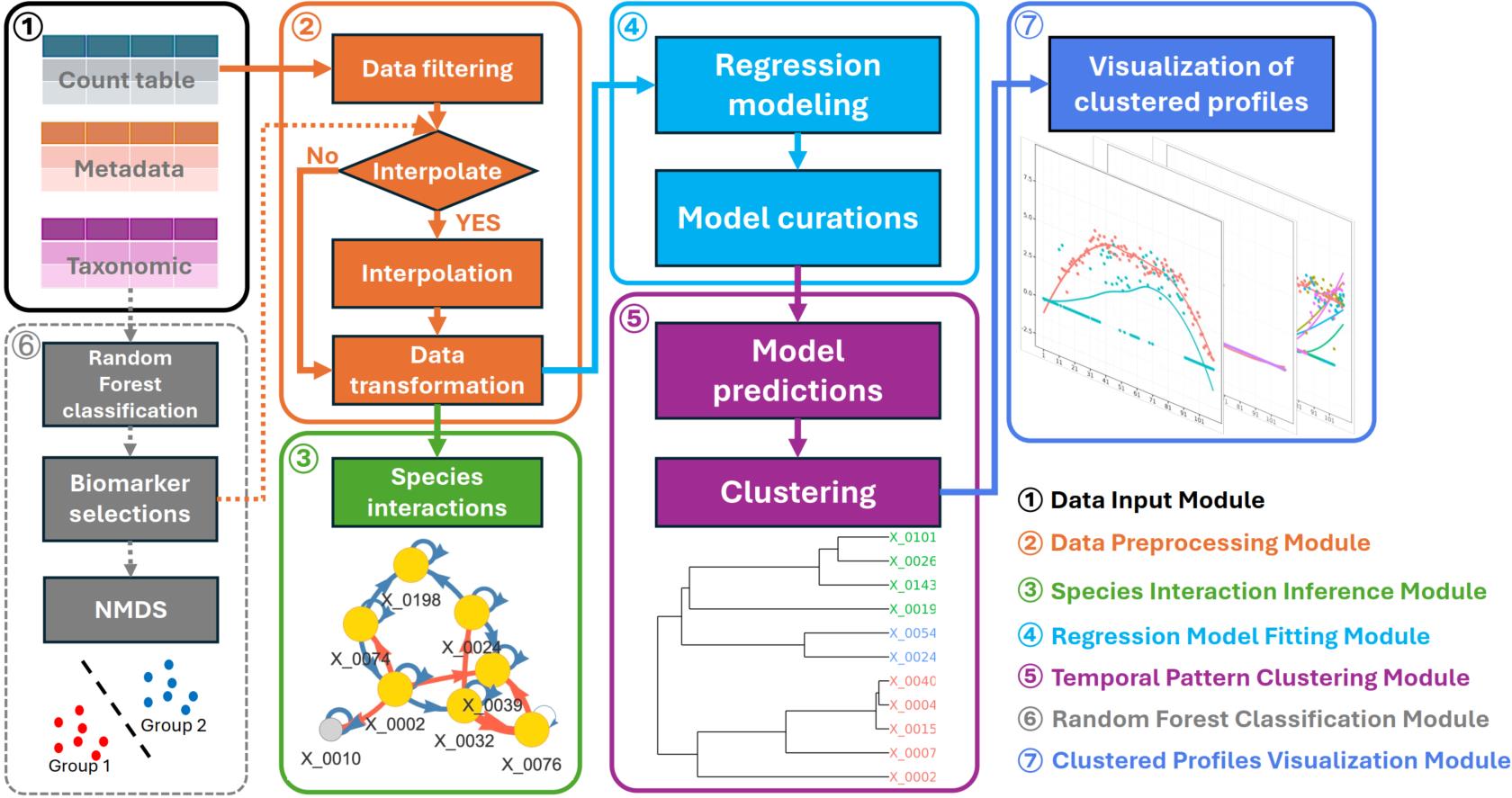
MicrobTiSDA is a user-friendly and flexible R package specifically designed for longitudinal microbiome data analysis. By integrating process-oriented functional modules—including data input, data preprocessing, interspecies interaction inference, natural spline regression modeling, temporal pattern clustering, and random forest classification—MicrobTiSDA enables users to efficiently and systematically infer reliable interspecies interactions and accurately characterize microbial abundance dynamics from time-series data. The package is compatible with diverse experimental designs and provides comprehensive, intuitive visualizations, thereby substantially enhancing both the efficiency of longitudinal microbiome data analysis and the depth of biological interpretation.
Weizmannia coagulans XY2 Mitigates Copper Neurotoxicity via Gut–Brain Axis Modulation of Tryptophan Metabolism and Oxidative-Inflammatory Crosstalk
- 18 November 2025
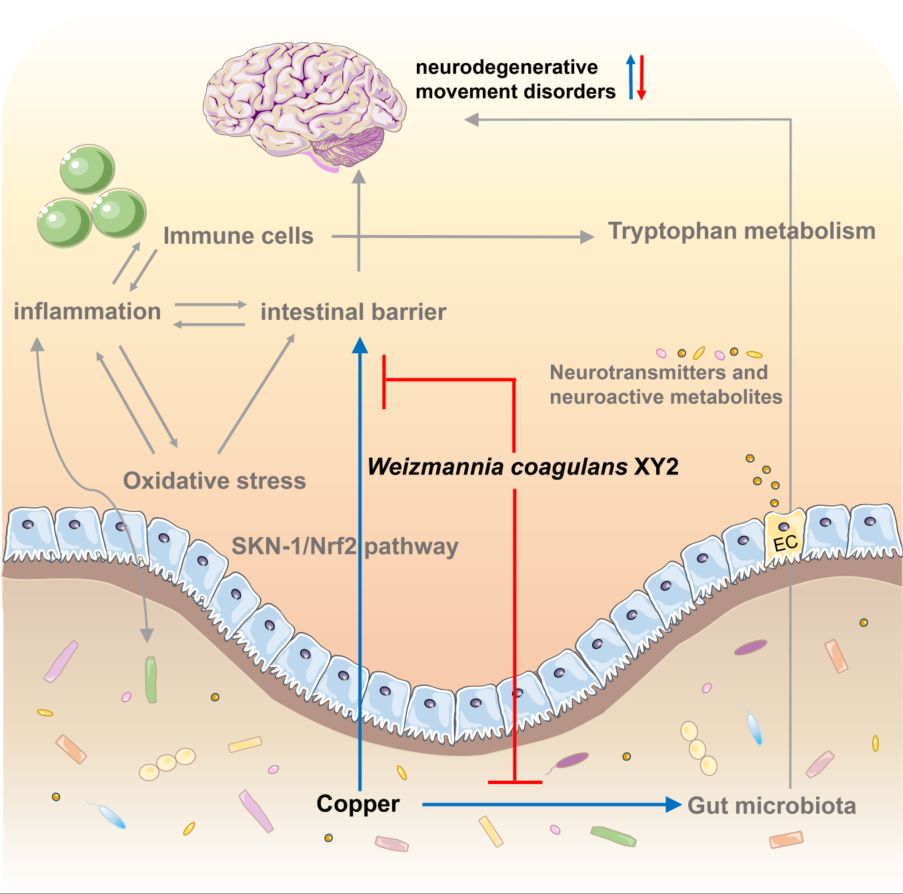
Copper interferes with tryptophan metabolism and 5-HT levels by modulating intestinal flora. Intestinal barrier breakdown and inflammatory response trigger nerve damage under copper exposure. W. coagulans XY2 alleviates copper-induced neurotoxicity by targeting a multi-dimensional “tryptophan metabolism-antioxidant defense-gut-brain axis” network.
CellOntologyMapper: Consensus mapping of cell type annotation
- 06 November 2025

CellOntologyMapper automates the standardization of cell type annotations in single-cell RNA-seq by mapping user-defined names to the Cell Ontology (CL) and Cell Taxonomy (CT). The framework constructs a comprehensive embedding database of 19,381 curated cell types using sentence transformers, large language models, and synonym expansion. User inputs, such as abbreviations (e.g., “dNK”), are processed through embedding similarity ranking and LLM amplification to resolve ambiguous or context-dependent labels. The system outputs standardized ontology terms with high accuracy (0.835 for CL, 0.911 for CT). Implemented as a Python package within the OmicVerse ecosystem and available via a web interface, CellOntologyMapper enables reproducible, large-scale single-cell meta-analysis by unifying diverse nomenclatures across studies, tissues, species, and disease states.
Host–microbiota interaction drives 5-hydroxyindole-3-acetic acid production to promote linear growth in infant mice
- 23 October 2025
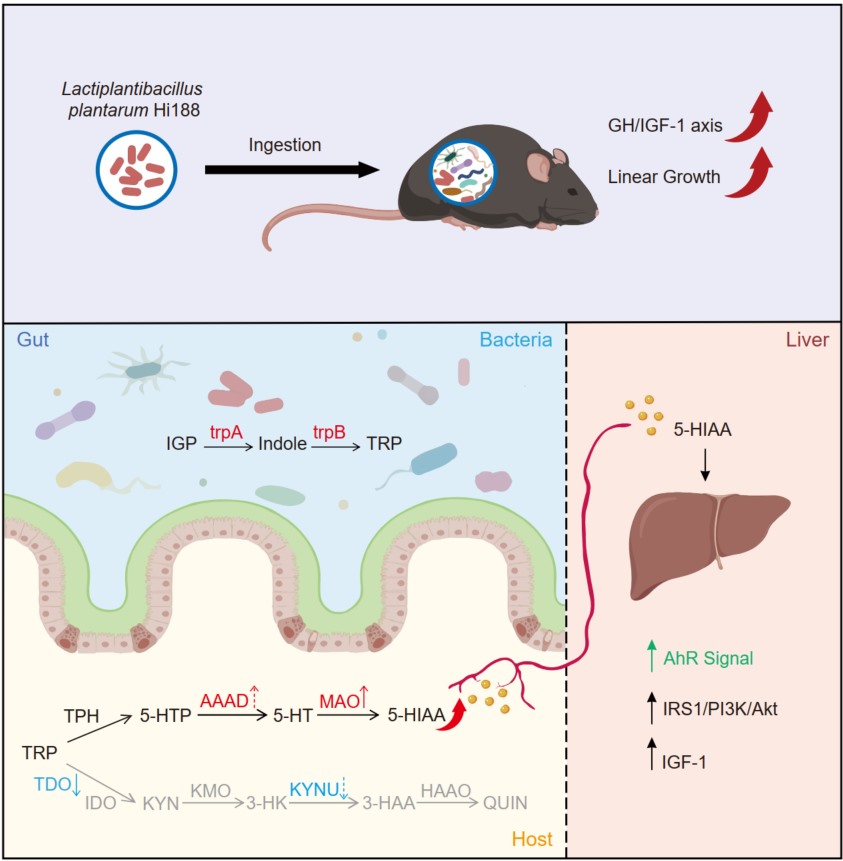
This study discovered that Lactiplantibacillus plantarum Hi188 promoted linear growth in postweaning mice. Transcriptomic analysis, untargeted metabolomics, and in vitro experiments showed that the elevated levels of 5-hydroxyindole-3-acetic acid (5-HIAA) activated the hepatic aryl hydrocarbon receptor (AhR) and subsequently promoted insulin-like growth factor-1 (IGF-1) production via the IRS1/PI3K/Akt pathway. Integrated metagenomics and reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis of host colonic genes revealed that 5-HIAA was cooperatively produced by the gut microbiota and the host. These findings offered novel perspectives on the therapeutic potential of microbiota-targeted interventions for growth regulation.
Rapid reconstruction of multidrug-resistant bacterial genomes using CycloneSEQ nanopore sequencing
- 26 October 2025
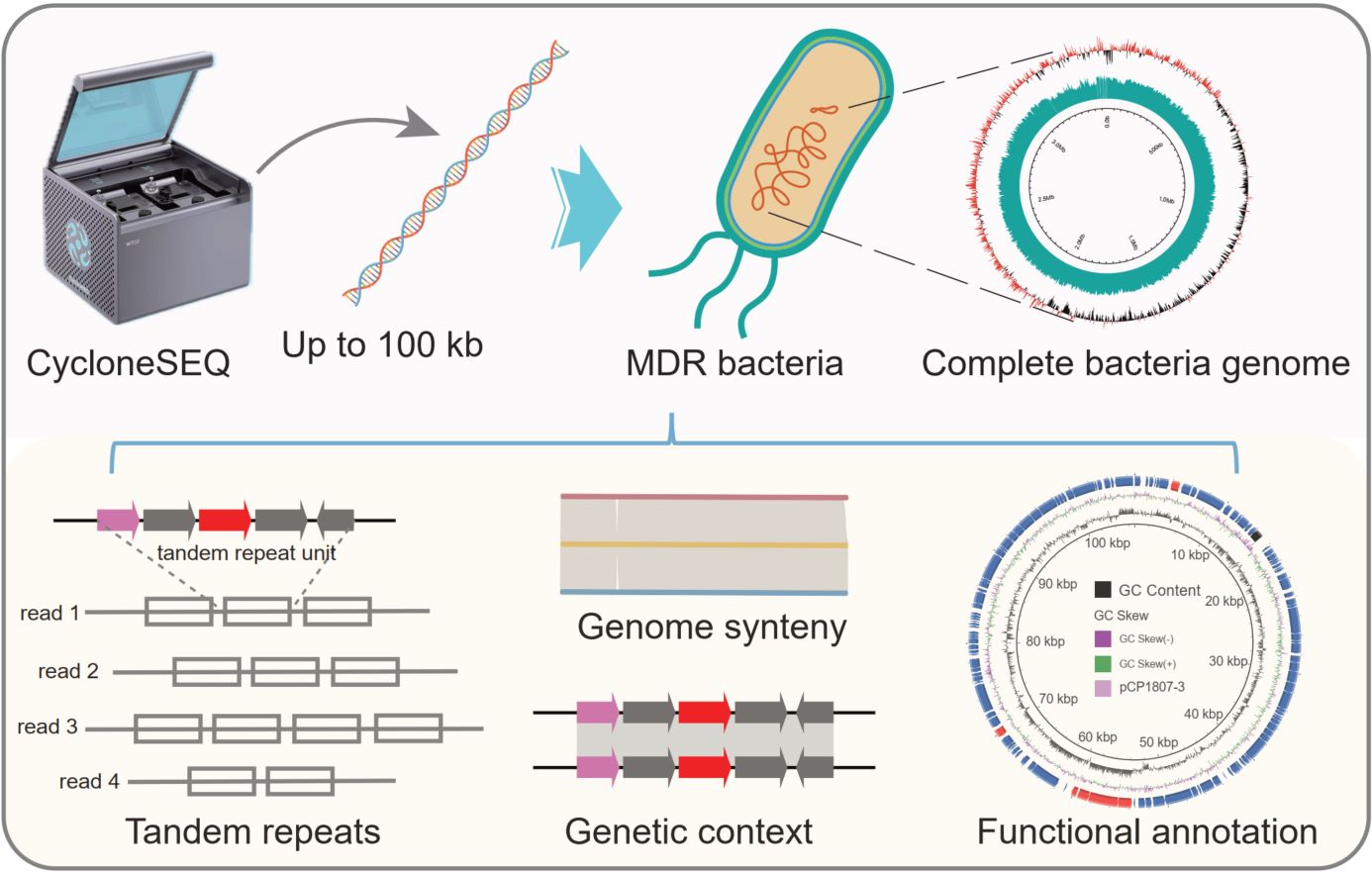
Multidrug-resistant (MDR) bacteria commonly harbor highly complex genomes enriched with mobile elements and resistance islands, which are challenging to resolve using short-read sequencing. Here, we evaluated CycloneSEQ, a newly developed Chinese nanopore sequencing platform, for MDR genome reconstruction. Long-read-only assemblies generated near-complete genomes, while hybrid assemblies achieved accuracy exceeding 99.99%. CycloneSEQ effectively resolved complex structures, including tandem repeats within ARG-bearing regions. Notably, updated sequencing chemistry improved single-read accuracy to 96.2% and yielded assembled genomes with 99.98% accuracy. These results demonstrate that CycloneSEQ enables the generation of complete and highly accurate bacterial genomes, highlighting its potential for antimicrobial resistance research and clinical surveillance.
Convergent evolution of metabolic functions: Evidence from the gut microbiomes of humans and dogs
- 21 October 2025
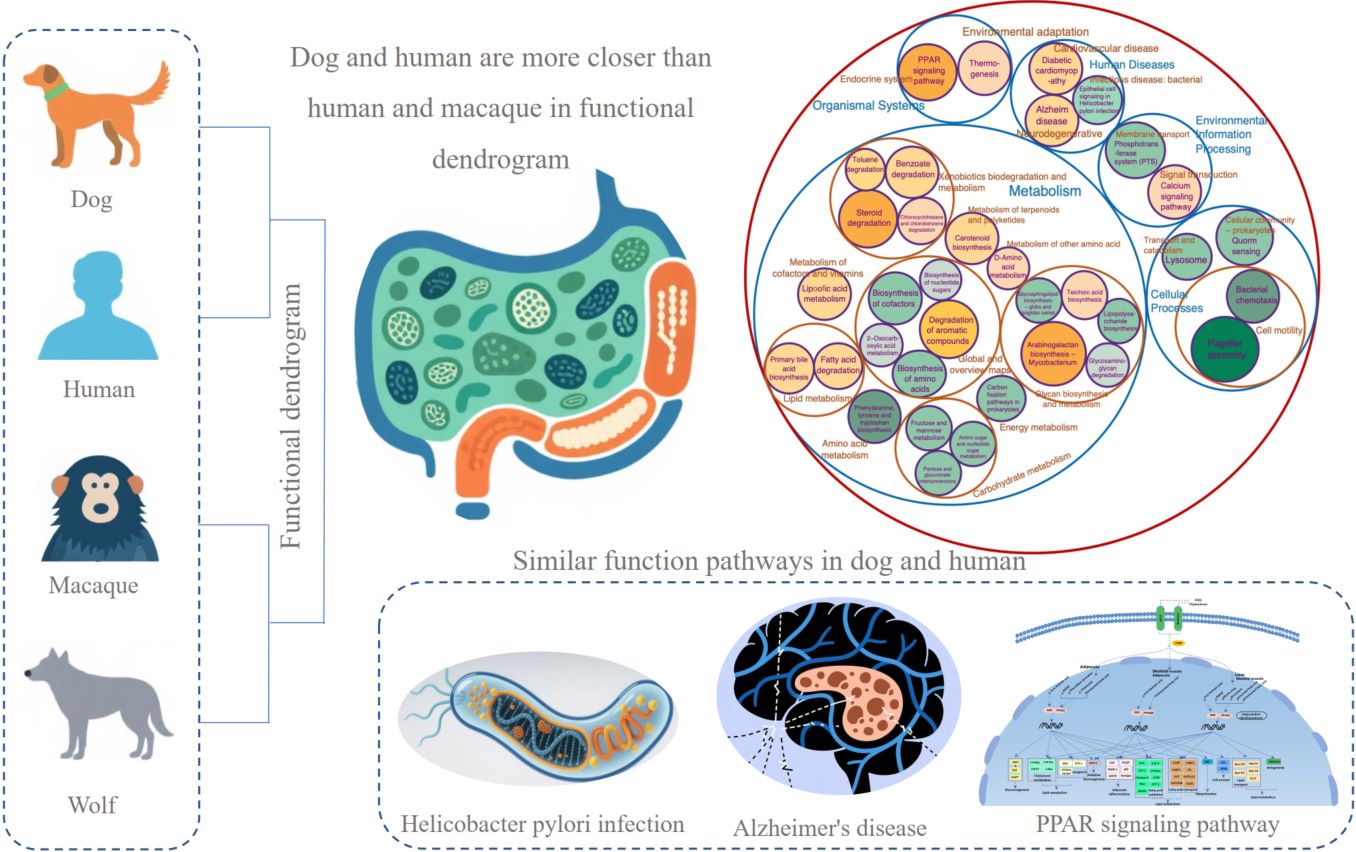
Dogs have undergone convergent evolution in their genome with humans in long-term co-living. Studies have shown that the gut microbiomes of dogs exhibit greater similarity to those of humans compared to mice and pigs. In our study, we first compared the gut microbiomes of macaques, three dog groups, and two wolf groups to those in humans. Results showed that gut microbiomes in dogs have undergone convergent evolution with those in humans. Functional enrichment analysis revealed that dogs exhibit the strongest carbohydrate metabolism ability compared to other hosts including starch and sucrose metabolism. Working dogs and pet dogs showed distinct functional specialization in metabolism and human disease. We suggest that dogs could serve as a potential model for research related to human gut microbial responses to human health, disease, and environmental changes.
Metagenomic mining reveals novel Cas12 subtypes and their evolutionary diversification
- 07 October 2025
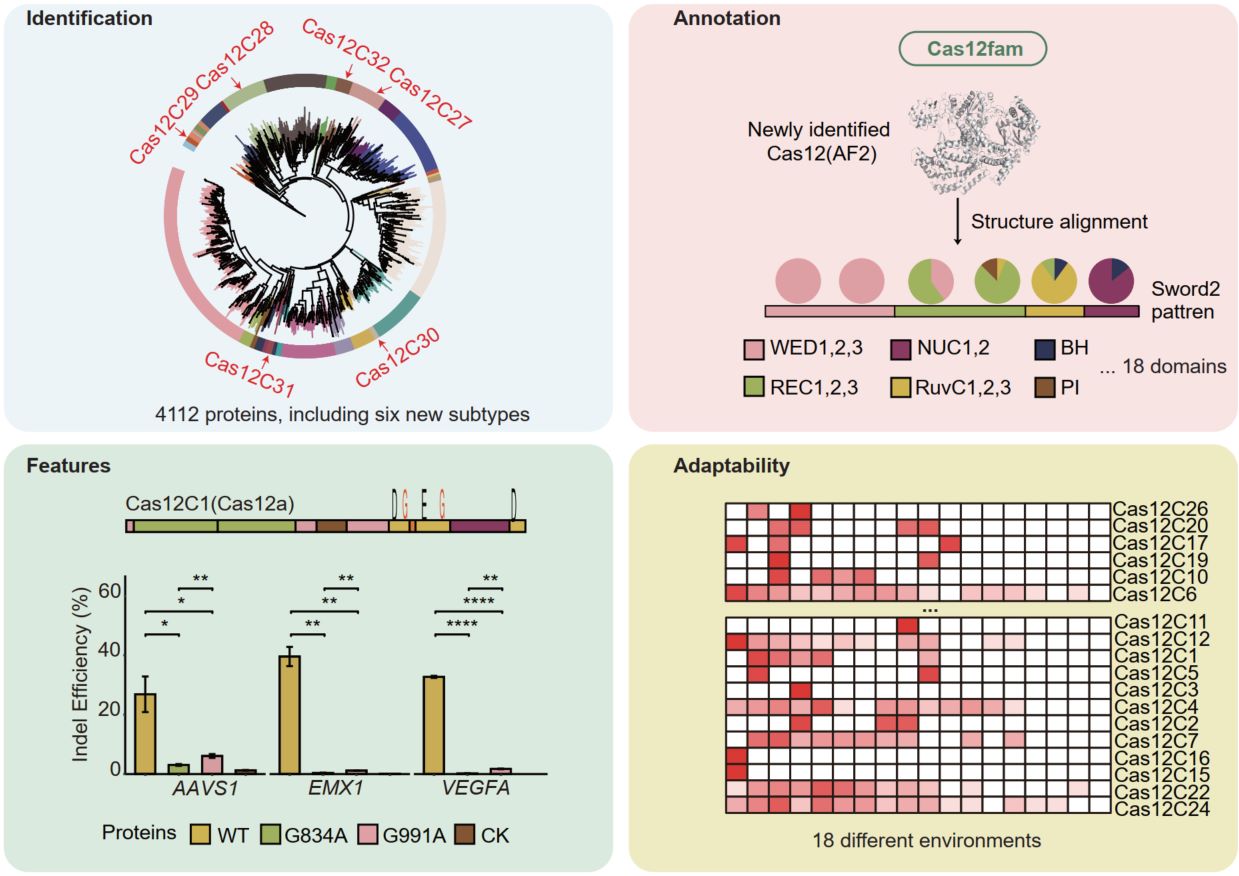
We identified 4112 Cas12 Proteins and 6 new Cas12 subtypes, revealed their significant diversity in N-terminal regions, repeat sequences, and sequences motifs. We developed an AI-driven algorithm, Cas12fam, which allows precise annotation of 18 distinct domains in Cas12 proteins. Through detailed structural analysis, we identified two novel glycine residues in the RuvC-like nuclease domains (RuvC-I, RuvC-II), which were experimentally validated to play a critical role in the DNA-cleavage activity of Cas12. Our analyses revealed that Cas12 proteins are enriched in specific environments, with significant variation in their abundance across ecological niches.
Early matrine intervention of gut microbiota for type 2 diabetes prevention
- 05 August 2025
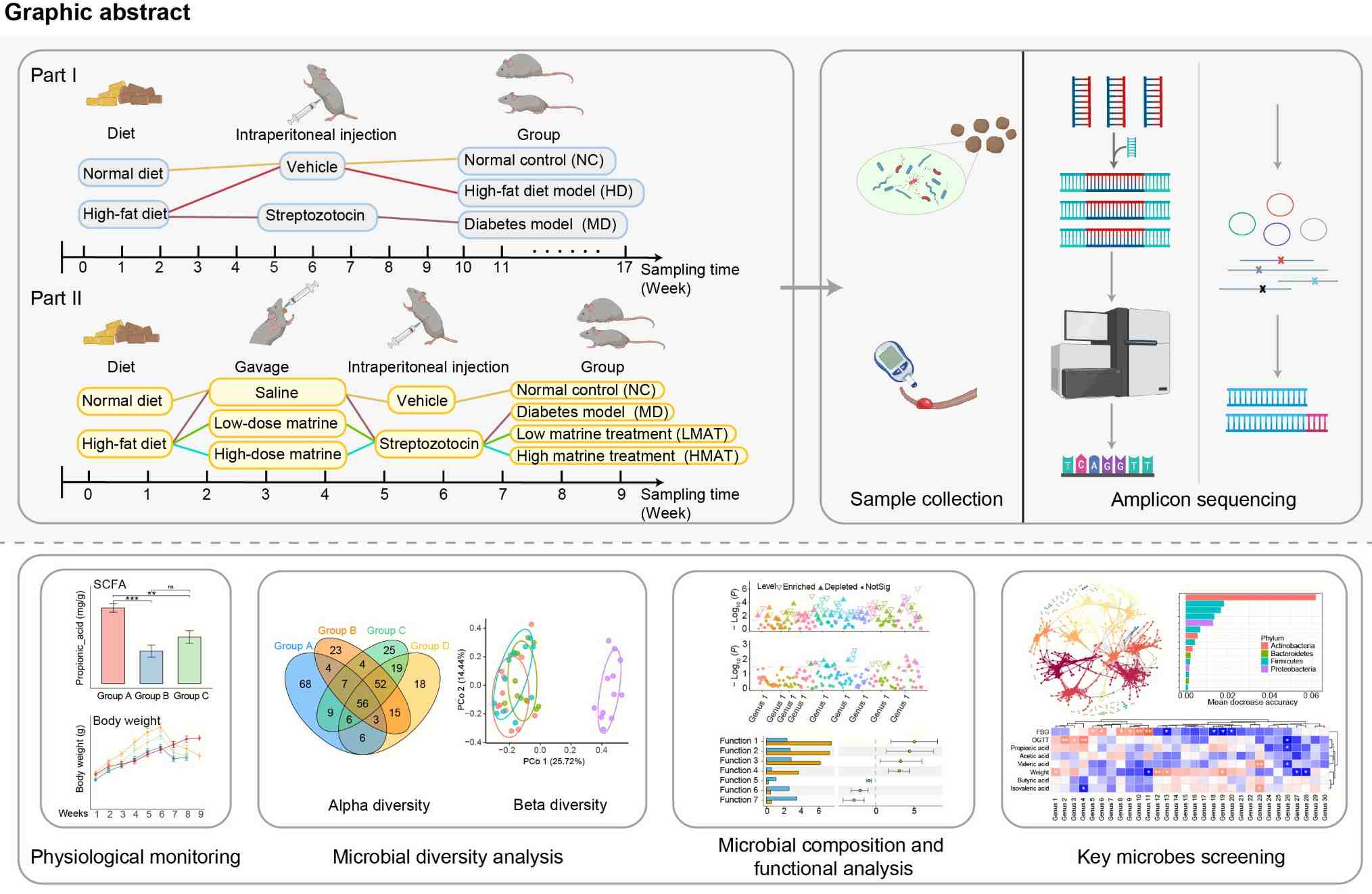
How matrine influences gut microbiota imbalance to prevent the progression of diabetes remains unclear. We conduct experiments using mice to simulate the stages of diabetes development and matrine intervention. Combined with amplicon sequencing, we find that the gut microbiota of diabetic mice continuously changes with the progression of the disease. Furthermore, differences in microbial community composition and function can distinguish the matrine intervention group from other groups. Muribaculum is generally enriched in the intervention group, while Lachnospiraceae_incertae_sedis is predominantly enriched in the diabetes group. These microbes may become new targets for diabetes intervention, providing new insights into diabetes treatment.
Impact of dietary live microbes and nondietary prebiotic/probiotic intake on osteoarthritis and rheumatoid arthritis development: Stratified findings from NHANES data
- 17 August 2024
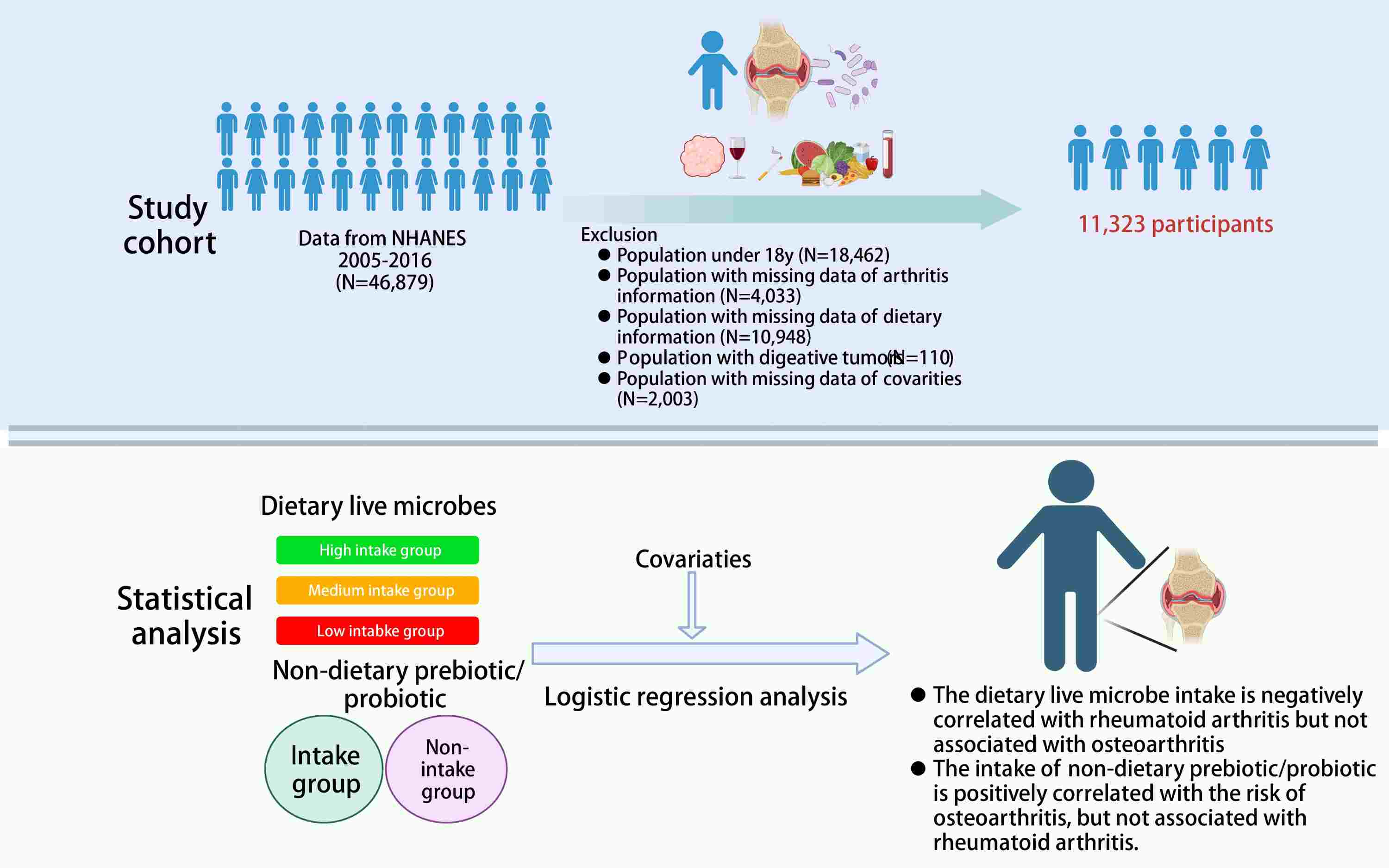
We selected participants from the NHANES database from 2005 to 2016 for this cross-sectional analysis. Logistic regression and other analytical methods were utilized to analyze the relationship between the intake of dietary live microbes and nondietary prebiotic/probiotic and the prevalence of osteoarthritis (OA) and rheumatoid arthritis (RA). The findings demonstrated a direct relationship between the consumption of nondietary prebiotic/probiotic and the prevalence of developing OA, whereas a greater consumption of dietary live microbes is associated with a lower occurrence of RA. (Graphical abstract was created with BioRender.com).
Cohort profile: A longitudinal multi-omics cohort of patients with acute coronary syndrome
- 03 July 2024
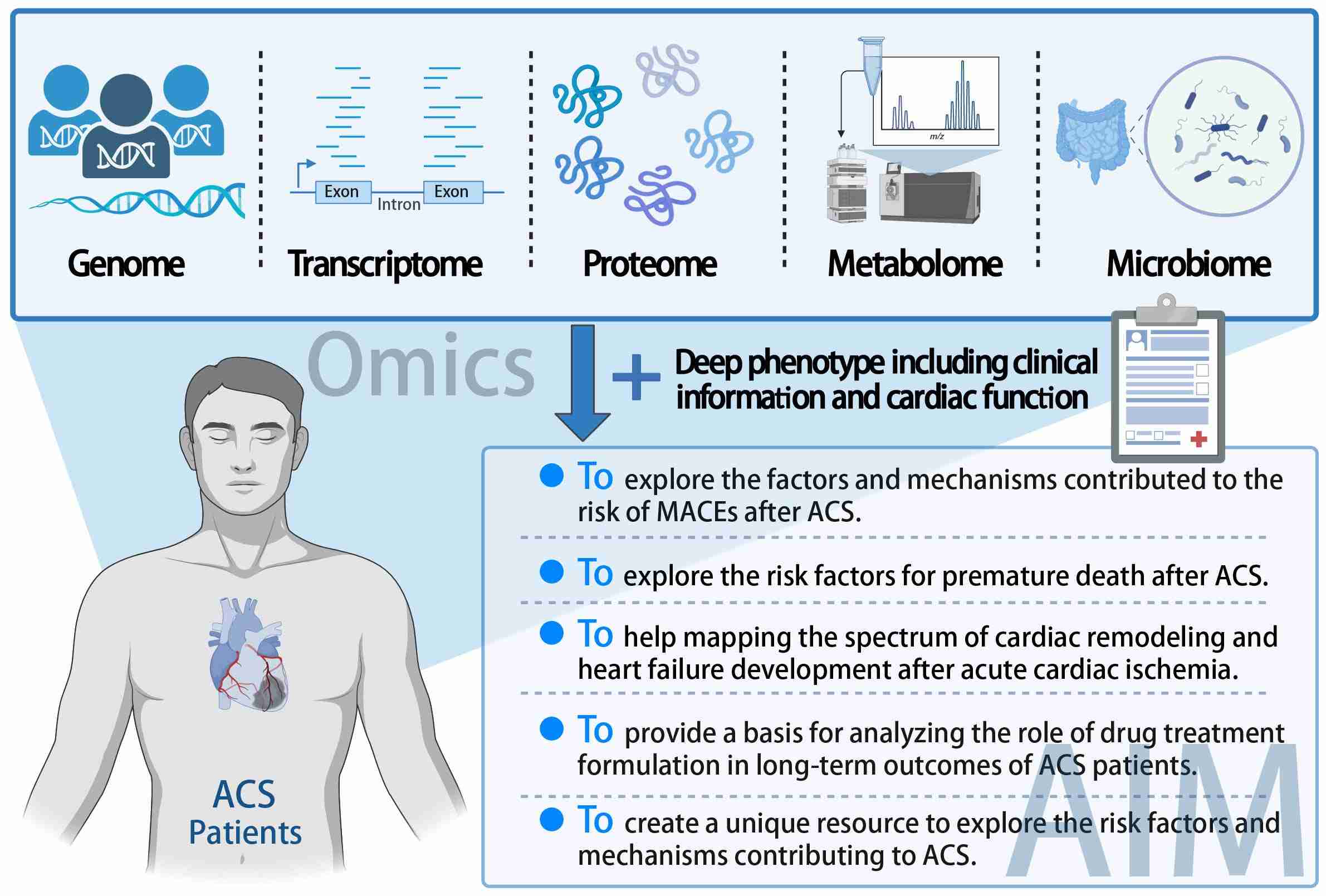
The longitudinal multi-omics cohort of patients with acute coronary syndrome (LM-ACS) is designed as a real-world prospective cohort of patients with ACS requiring coronary angiography. This study aims to enroll 50,000 participants, with a thorough collection of phenotypic data and multi-omics analyses performed on biological samples. Additionally, long-term follow-up will be conducted to track the incidence of major adverse cardiovascular events (MACEs) over the participants' lifetimes. For this purpose, three follow-up scenarios have been established for ACS patients, differentiated based on whether the patients had acute myocardial infarction (AMI) or unstable angina (UA), and whether they underwent percutaneous coronary intervention (PCI) surgery. The LM-ACS cohort seeks to create a unique resource to advance our understanding of the etiology and clinical outcomes of ACS.
The active effect of Rhizophagus irregularis inoculants on maize endophytic bacteria community
- 17 August 2024
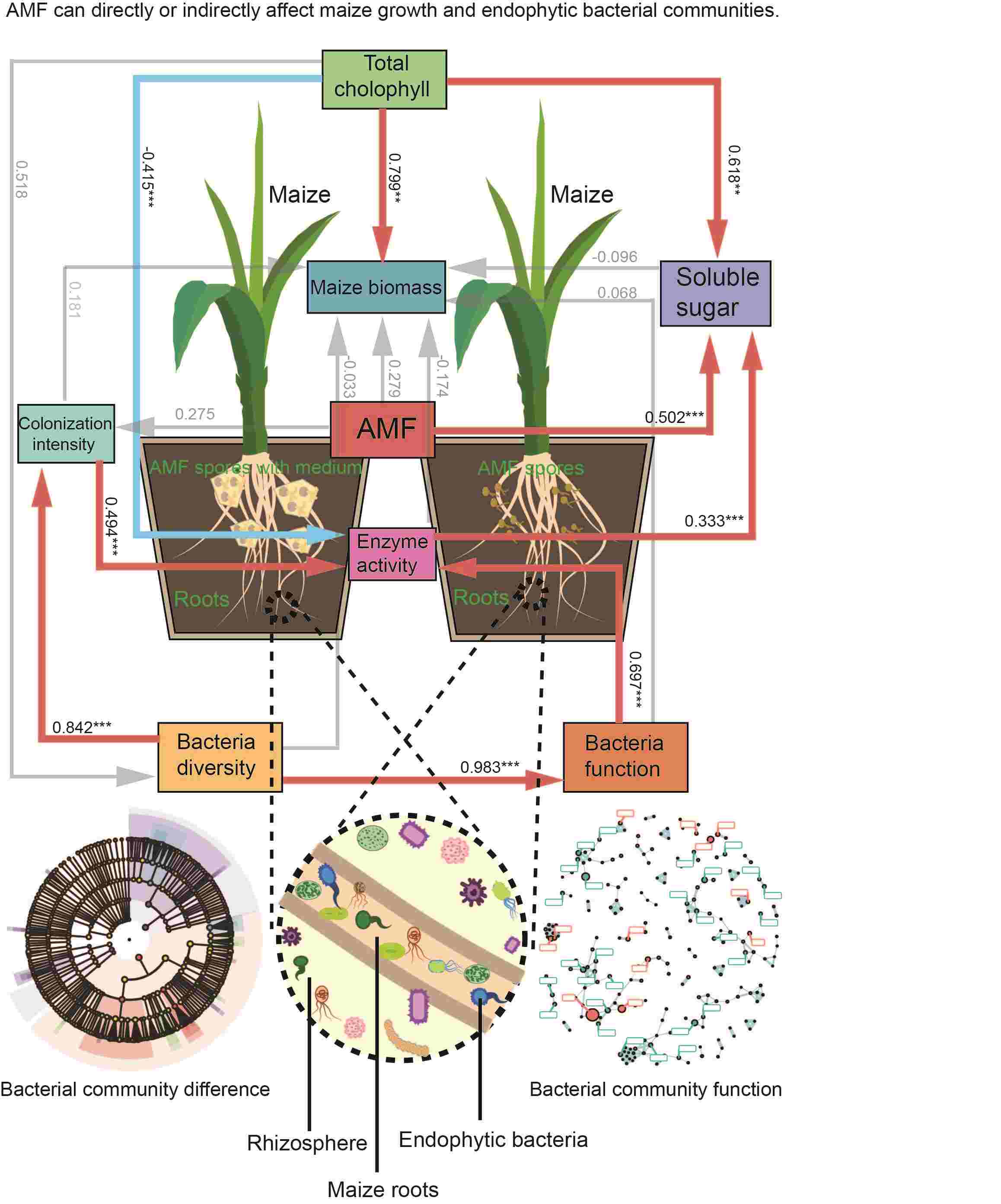
Little has been reported on the effect of arbuscular mycorrhizal fungi (AMF) inoculants from in vitro dual culture system on the growth of maize and its endophytic microbial community. Our results suggest that AMF inoculants play an important role in influencing maize growth and diversity of endophytic bacterial communities. AMF inoculants significantly promote maize growth, especially AMF inoculants with modified Strullu-Romand (MSR) medium. These inoculants also significantly increased the diversity of endophytic microbial communities, especially the abundance of beneficial bacterial flora, thus positively affecting maize growth. This study reveals the utility of AMF inoculants from in vitro dual culture system, which provides a basis for the development of environmentally friendly inoculants.
A glimpse into the future: Integrating artificial intelligence for precision HER2-positive breast cancer management
- 02 August 2024
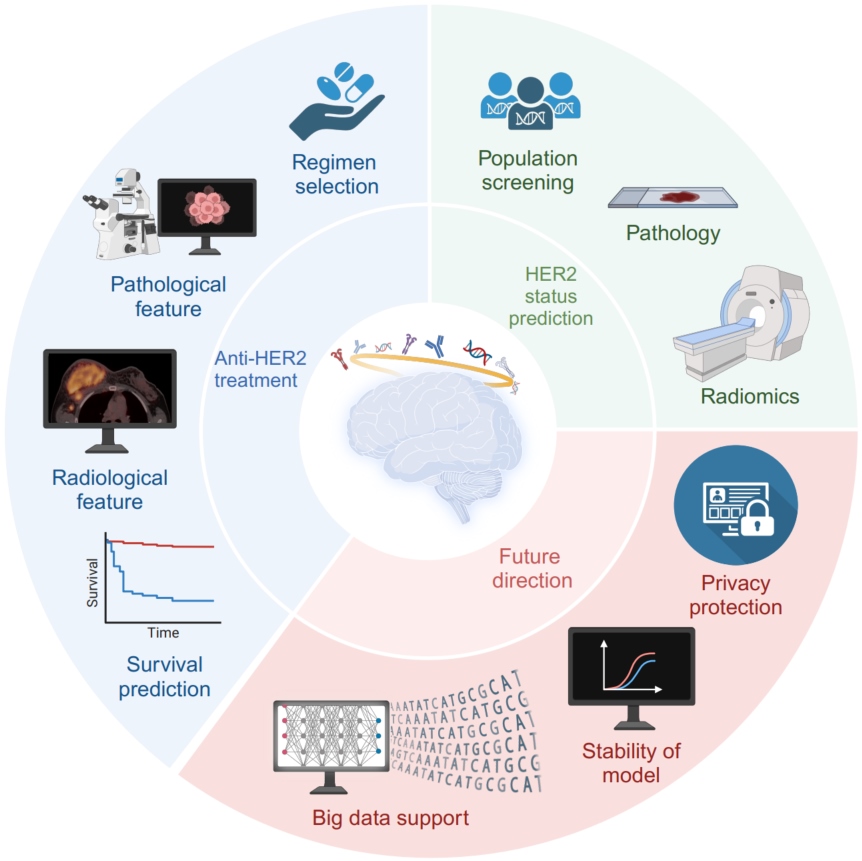
Breast cancer (BC), specifically HER2-positives subtype, has a poor prognosis. Nevertheless, the development of anti-HER2 therapy yielded satisfactory outcomes. Therefore, evaluating patient HER2 status and ascertaining responsiveness to anti-HER2 therapy is crucial. The advent of deep learning has propelled the artificial intelligence (AI) revolution, leading to an increased applicability of AI in predictive models. In the field of medicine, AI is an emerging modality that is gaining momentum for facilitating cancer diagnosis and treatment, particularly in the effective management of breast cancer. This study aims to provide a comprehensive review of current diagnostic and predictive models that utilize data obtained from histopathological slides, radiomics, and HER2 binding sites. Advancements and practical applications of these models were also evaluated. Additionally, we examined existing obstacles that AI encounters for anti-HER2 therapy. We also proposed future directions for integrating AI in assessing and managing anti-HER2 therapy. The findings of this study offer valuable insights into the evaluation of AI-based anti-HER2 therapy, emphasizing key concepts and obstacles that, if addressed, could facilitate the integration of AI-assisted anti-HER2 therapy. The integration of AI has the potential to enhance the precision and customization of screening and treatment protocols for HER2+ breast cancer.
TCfinder: Robust tumor cell discrimination in scRNA-seq based on gene pathway activity
- 07 August 2024
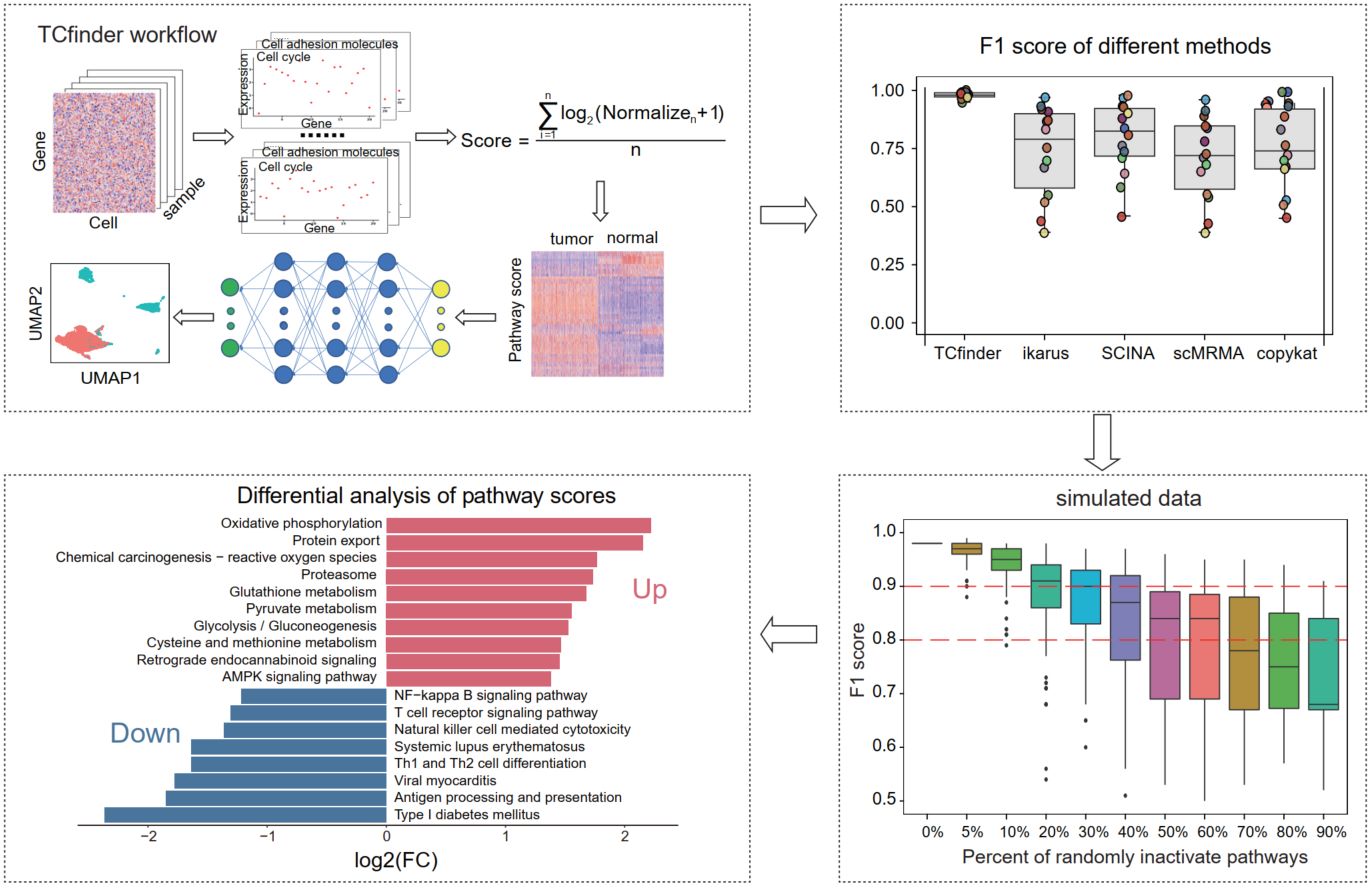
TCfinder is a tumor cell identification tool, based on pathway activity and deep neural network (DNN). Across different platforms of scRNA-seq datasets, TCfinder demonstrates robust identification efficiency. It outperforms existing tumor cell identification tools and performs under sparse data. TCfinder is freely available as an R package at: https://github.com/XSLiuLab/TCfinder.
Role of soil microbes in enhancing crop heterosis
- 01 August 2024
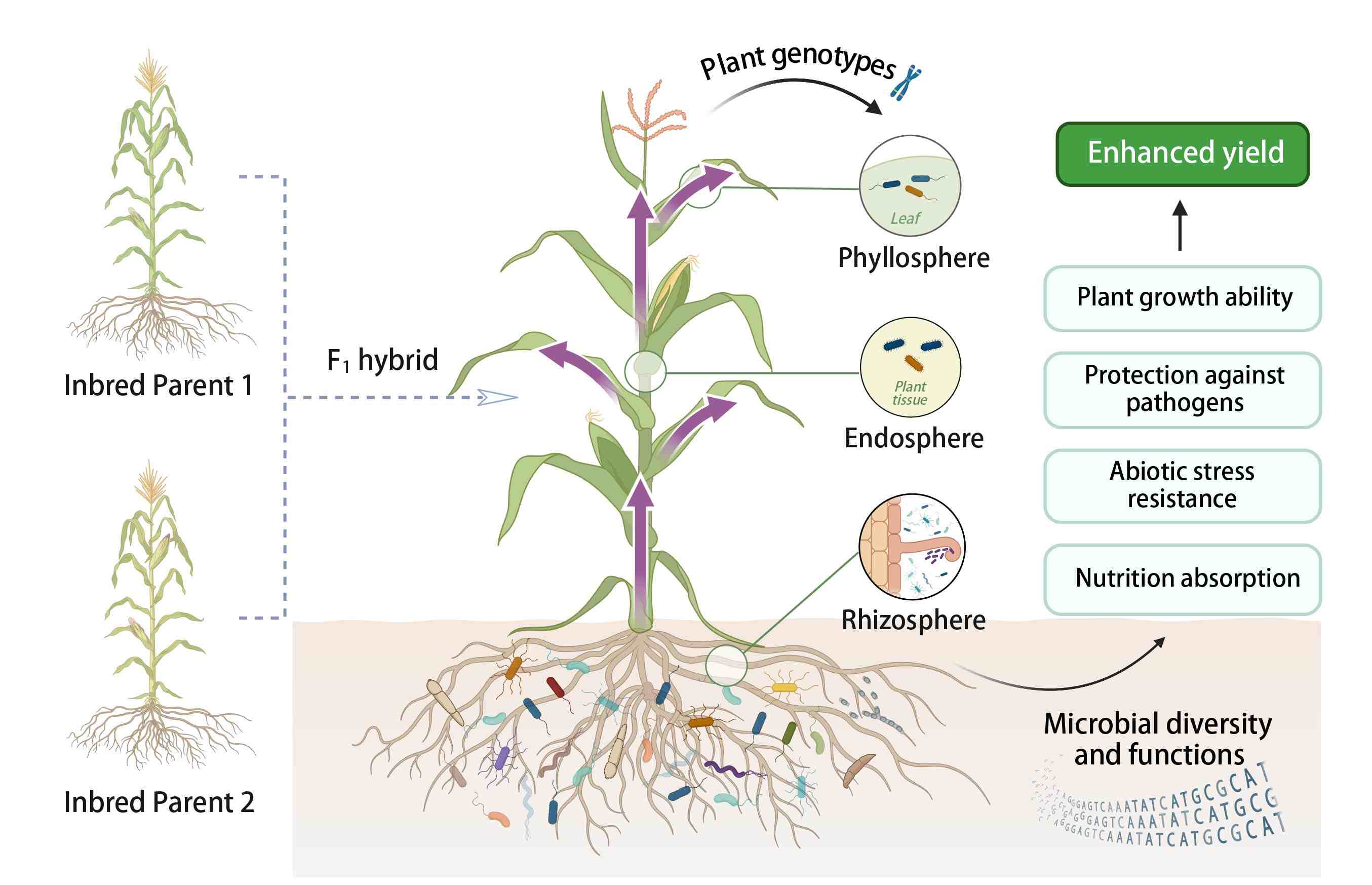
Heterosis, or hybrid vigor, is characterized by the enhanced performance of F1 progeny in terms of yield, biomass, and environmental adaptation compared to their parental lines. Recent studies underscore the significant influence of soil microbes on heterosis, revealing that plant genotypes shape microbial communities which, in turn, have the potential to support plant growth through complex host-microbe interactions. The deeper insight into microbial roles suggests innovative ways to boost crop performance and sustainability by managing the plant microbiome to further enhance heterosis
Emerging nanomedicine strategies for hepatocellular carcinoma therapy
- 04 July 2024
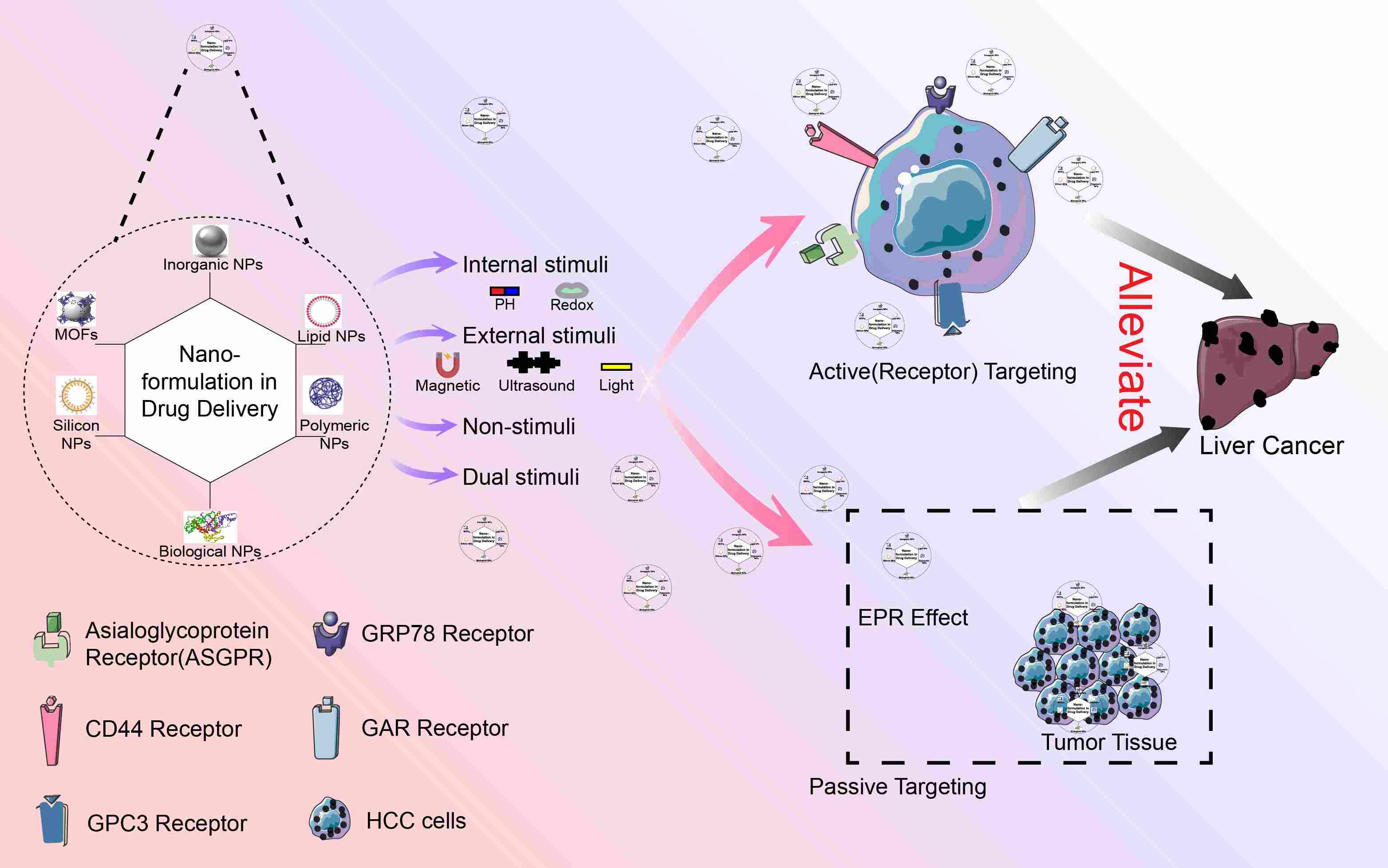
Hepatocellular carcinoma (HCC) is a prevalent malignant tumor with a range of risk factors, including viral infections, alcoholic liver disease, exposure to fungal toxins, obesity, and type 2 diabetes. Despite advancements in early detection and treatment, HCC continues to exhibit a high rate of recurrence. Patients in advanced stages have a poor prognosis, and the survival rate after cancer metastasis is notably low. Consequently, there exists an urgent necessity for novel treatment strategies. Nanomedicine possesses superior attributes, such as targeted administration and responsive drug release within the tumor microenvironment. These systems demonstrate potential in HCC treatment by facilitating the transportation of diverse therapeutic drugs. This comprehensive review aims to delve into the application of nanoparticle drug delivery systems in the management of liver tumors, with particular emphasis on formulation, targeted strategies specific to liver tumors, and various methods of drug release. By offering insights into the utilization of nanoparticle drug delivery systems in the realm of liver tumor treatment, this review endeavors to assist readers in exploring their vast potential.
Ocular microbiota types and longitudinal microbiota alterations in patients with chronic dacryocystitis with and without antibiotic pretreatment
- 09 July 2024
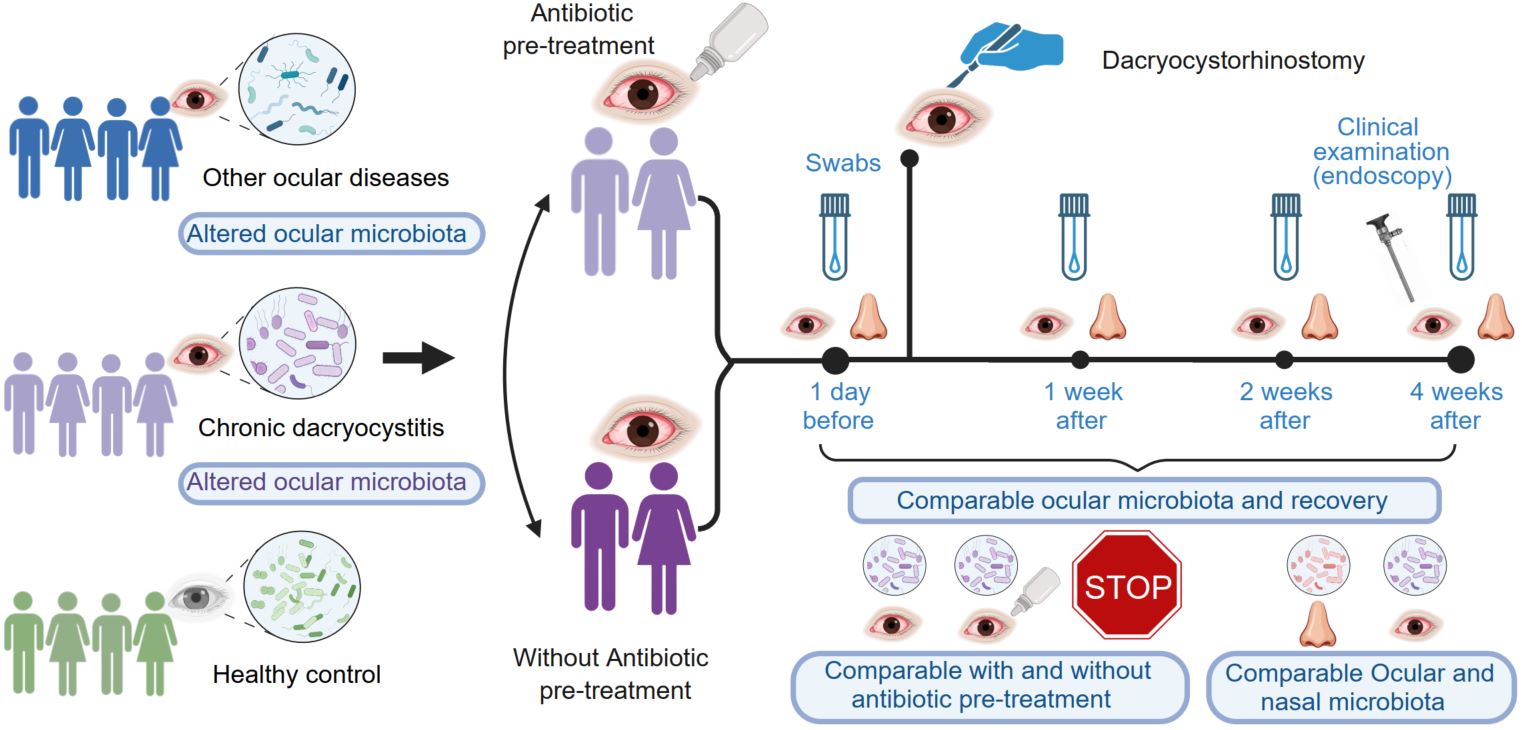
Antibiotic pretreatment is routine for chronic dacryocystitis (DC) patients. Herein, the longitudinal effects of antibiotic pretreatment before dacryocystorhinostomy for DC patients were evaluated. Conjunctival and nasal swabs were collected longitudinally from 33 DC patients with and without antibiotic pretreatment, both before dacryocystorhinostomy and at 1, 2, and 4 weeks postdacryocystorhinostomy. Additionally, conjunctival sac swabs were collected from 46 healthy volunteers and 14 other ocular diseases patients. Comparisons focused on ocular/nasal microbiota and recovery outcomes. Compared to healthy participants, DC patients without antibiotic pretreatment exhibited greater ocular microbiota diversity before dacryocystorhinostomy. Although clinical recovery rates were comparable, our results suggest that, after antibiotic pretreatment, the ocular microbiota richness and diversity, and the composition alteration tendency, significantly changed 4 weeks after surgery. This implies that the ocular microbiota was more disturbed in patients who underwent antibiotic pretreatment compared to those without such treatment. Furthermore, two types of ocular microbiota and three types of nasal microbiota were identified in ocular diseases. This study provides comprehensive data on the ocular and nasal microbiota in DC patients with and without antibiotic pretreatment, along with other ocular diseases. This finding suggested that antibiotic pretreatment may not be necessary before dacryocystorhinostomy for DC patients, especially for nonsevere cases.
Insights into the evolution and fruit color change-related genes of chromosome doubled sweet cherry from an updated complete T2T genome assembly
- 30 June 2024
Combined detection of inflammatory proteins is beneficial for diagnosing the papillary thyroid carcinoma and nodular goiter
- 02 July 2024
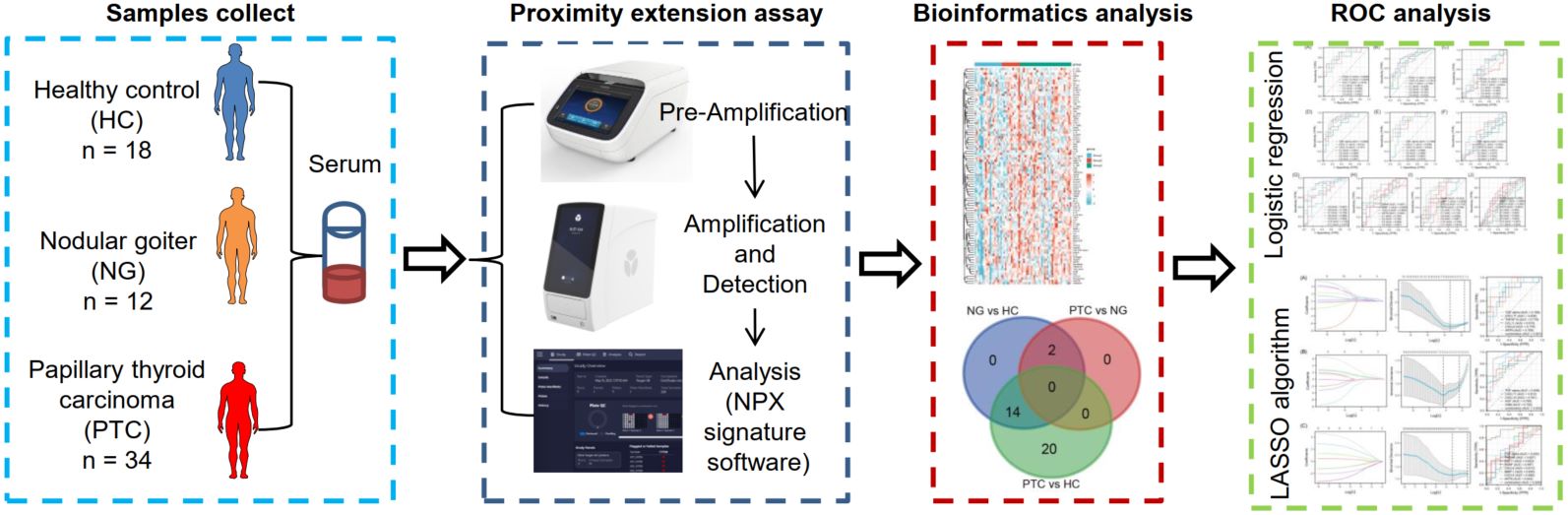
Fine-needle aspiration cytology and imaging examinations are commonly used diagnostic tools for papillary thyroid carcinoma (PTC). However, these methods have limitations. Inflammatory proteins have the potential to serve as diagnostic and prognostic markers, as well as treatment targets. The expression profile and diagnosis effect of inflammatory proteins in PTC are not well understood. Here, 18 healthy volunteers (as healthy control), 12 patients with nodular goiter, and 34 patients with PTC were collected to analyze serum inflammatory proteins by proximity extension assay. Receiver operating characteristic curve analysis was used to evaluate the diagnostic potential of differential expression of proteins via the area under the curve (AUC) analysis. A total of 36 differentially expressed inflammatory proteins were found among PTC, nodular goiter, and healthy control. The combination diagnosis derived from the logistic regression analysis exhibited promising diagnostic capabilities in distinguishing nodular goiter from healthy control (AUC = 0.88), distinguishing PTC from healthy control (AUC = 0.89), and distinguishing PTC from nodular goiter (AUC = 0.87). Whereas the combination diagnosis derived from the least absolute shrinkage and selection operator (LASSO) exhibited promising diagnostic capabilities in distinguishing nodular goiter from healthy control (AUC = 0.92), distinguishing PTC from healthy control (AUC = 0.93), and distinguishing PTC from nodular goiter (AUC = 0.93). Overall, this study offers potential biomarkers for distinguishing between PTC and nodular goiter in clinical practice. The combination derived from the LASSO algorithm outperforms logistic regression.
Developing glycoproteomics reveals the role of posttranslational glycosylation in the physiological and pathological processes of male reproduction
- 01 July 2024
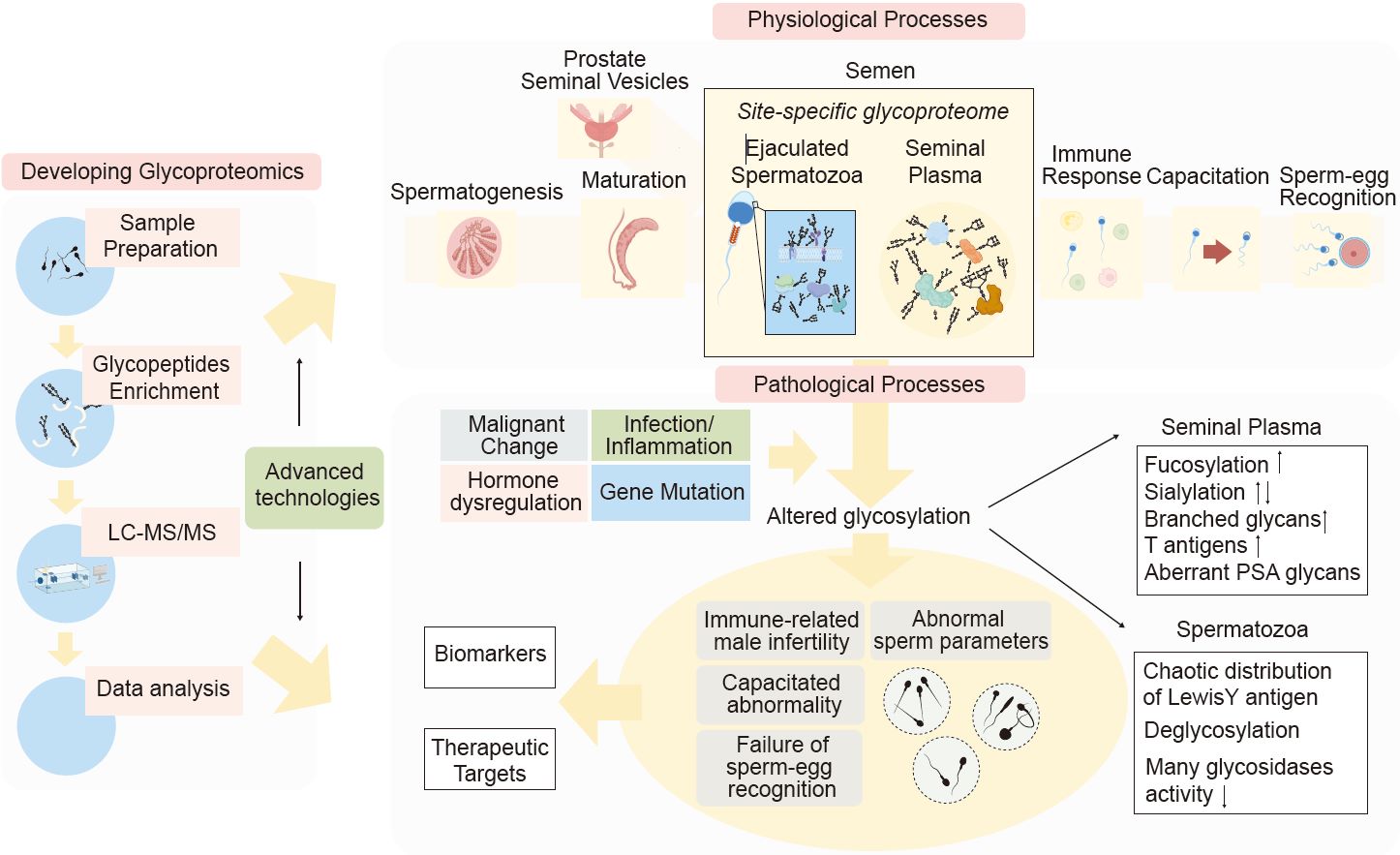
Glycosylation plays a pivotal role in the physiological and pathological processes of male reproduction. It impacts thousands of proteins and is actively ongoing through all stages of reproduction, including spermatogenesis, maturation, capacitation, and fertilization. However, our grasp on glycosylation within male reproductive processes remains limited, largely due to the technical hurdles. Recent advancements have seen the mapping of the glycoproteome of human semen, utilizing cutting-edge glycoproteomic technologies. This breakthrough lays the groundwork for in-depth research into the influence of glycosylation on male reproductive system and related disorders. Nevertheless, the field faces numerous challenges that necessitate further advancements in glycoproteomic methodologies. In this analysis, we evaluated the potential applications of advanced glycoproteomic techniques in the study of male reproduction and summarized the detailed profiling of the human semen glycome and glycoproteome. Our current understanding of glycosylation's role within the male reproductive system alongside recent progress in glycoproteomics may equip biologists with a comprehensive insight. Furthermore, this analysis brought together findings on abnormal glycosylation and its link to male reproductive disorders in the view of glycomics and glycoproteomics. It can facilitate the clinical application of glyco-related biomarkers and targets in the treatment of infertility.
A super pan-genome map provides genomic insights into evolution of diploid cotton species
- 27 June 2024
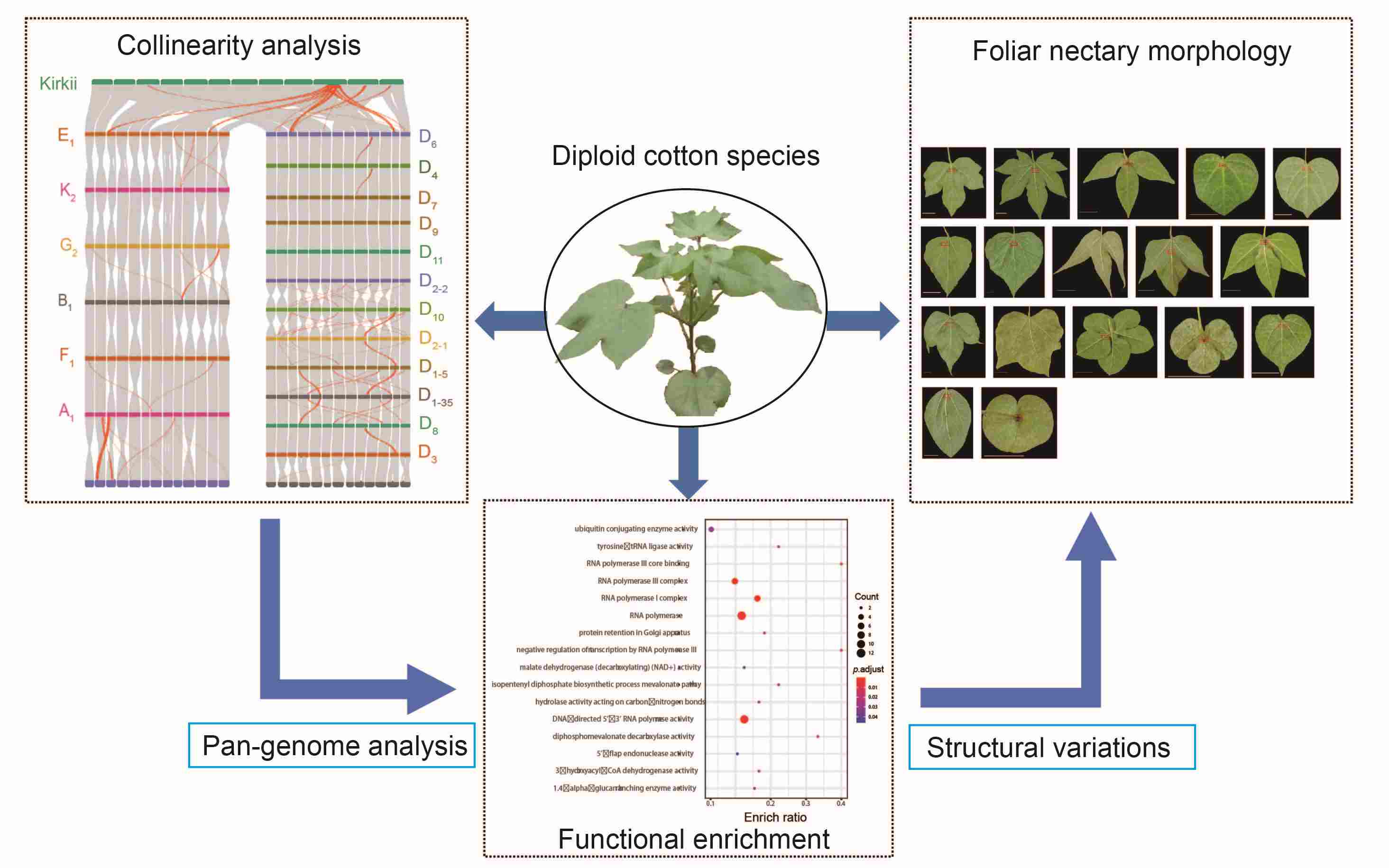
The 45 diploid cotton species identified worldwide exhibit remarkable morphological diversity. Modern cotton breeding is limited by incomplete understanding of the genetic variation in these species, suggesting a need for pan-genomic comprehensive analyses. In this study, a high-quality super pan-genome was built using 22 representative diploid cottons species and their adaptive evolution was investigated. The genomes of the twenty-two species yielded an average of 923,706 transposable elements (TEs) per assembly, with TE proportions ranging from 62.29% to 88.92%. The inferring ancestor genome structure (IAGS) showed that the D5 genome was closer to the ancestor, and the K2 genome accumulated more fissions and fusions. A gene-based super pan-genome identified 67,807 genes, including 22,384 core, 34,093 variable, and 11,330 specific genes. The structural variations (SVs) were unevenly distributed on the chromosomes, and 321 hotspot regions were detected, containing 90 genes associated with fiber initiation and/or elongation. During the eastward diffusion of diploid cotton, the genome size and structure experienced significant changes. We investigated the foliar nectary in 17 diploid cotton species, and identified a 444-bp deletion in the promoter sequence of GoNe that explained the lack of foliar nectary in G. gossypiodes (D6) and G. schwendimanii (D11). This pan-genome construction and comprehensive analysis for diploid cotton provided insight into dynamic genomic variation during diploid cotton expansion and can facilitate effective modern cotton breeding.
Long journey of 16S rRNA-amplicon sequencing toward cell-based functional bacterial microbiota characterization
- 11 July 2024
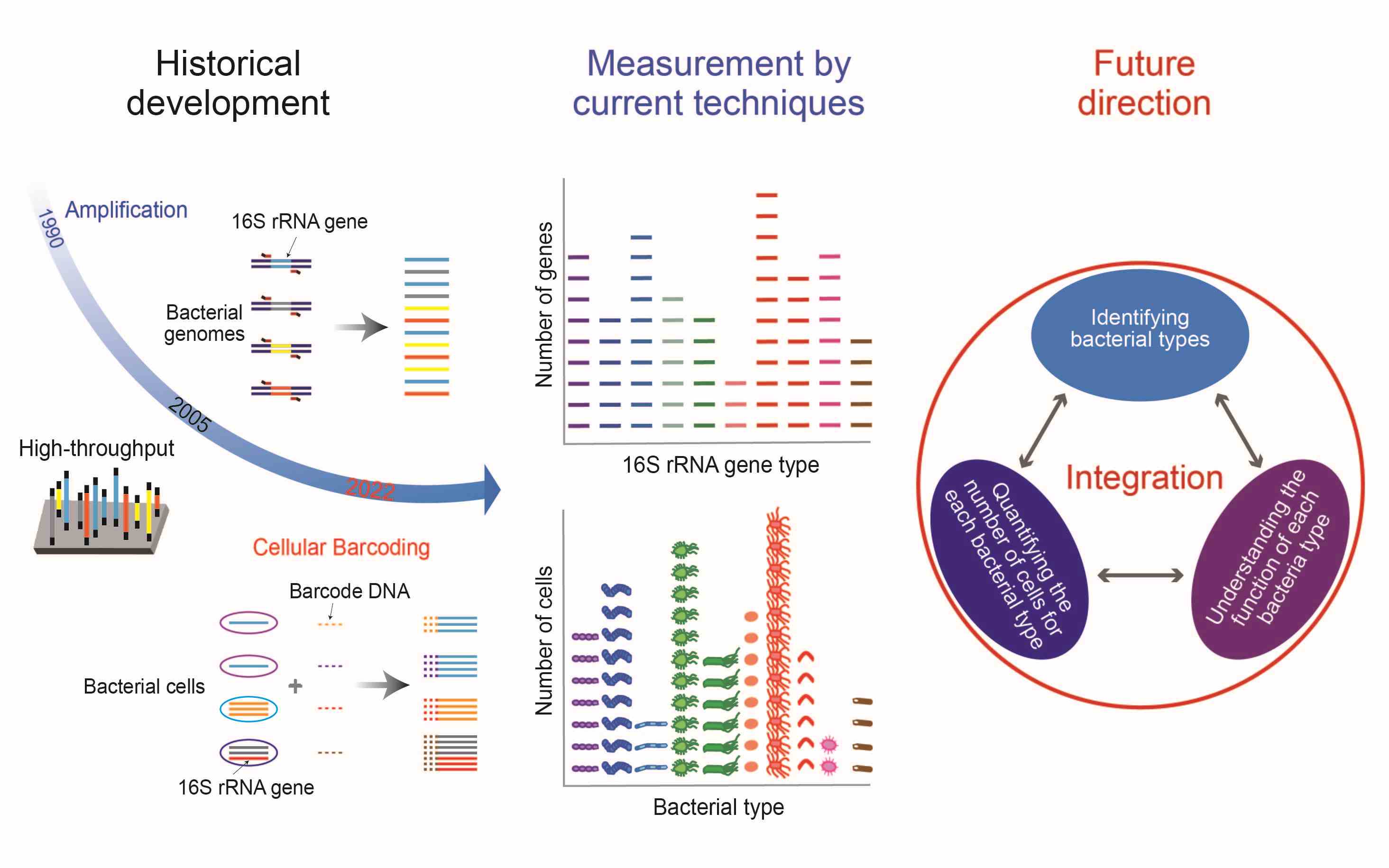
Bacteria often exist and function as a community, known as the bacterial microbiota, which consists of vast numbers of bacteria belonging to many bacterial species (taxa). Characterizing the bacterial microbiota needs high-throughput approaches that enable the identification and quantification of many bacterial cells, and such approaches have been under development for more than 30 years. In this review, we describe the history of high-throughput technologies based on 16S ribosomal RNA (rRNA) gene-amplicon sequencing for the characterization of bacterial microbiotas. Then, we summarize the features and applications of current 16S rRNA gene-amplicon sequencing approaches, including a recent achievement that enables the identification of individual cells with single-base accuracy for 16S rRNA genes and the quantification of many identified cells. Furthermore, we present the prospects for further technical development, including the combined use of high-throughput methods and other informative analyses, such as whole-genome sequencing in the common unit of the cell, which enables bacterial microbiota characterization based on both the number of cells and their functions.
STAGER checklist: Standardized testing and assessment guidelines for evaluating generative artificial intelligence reliability
- 02 July 2024
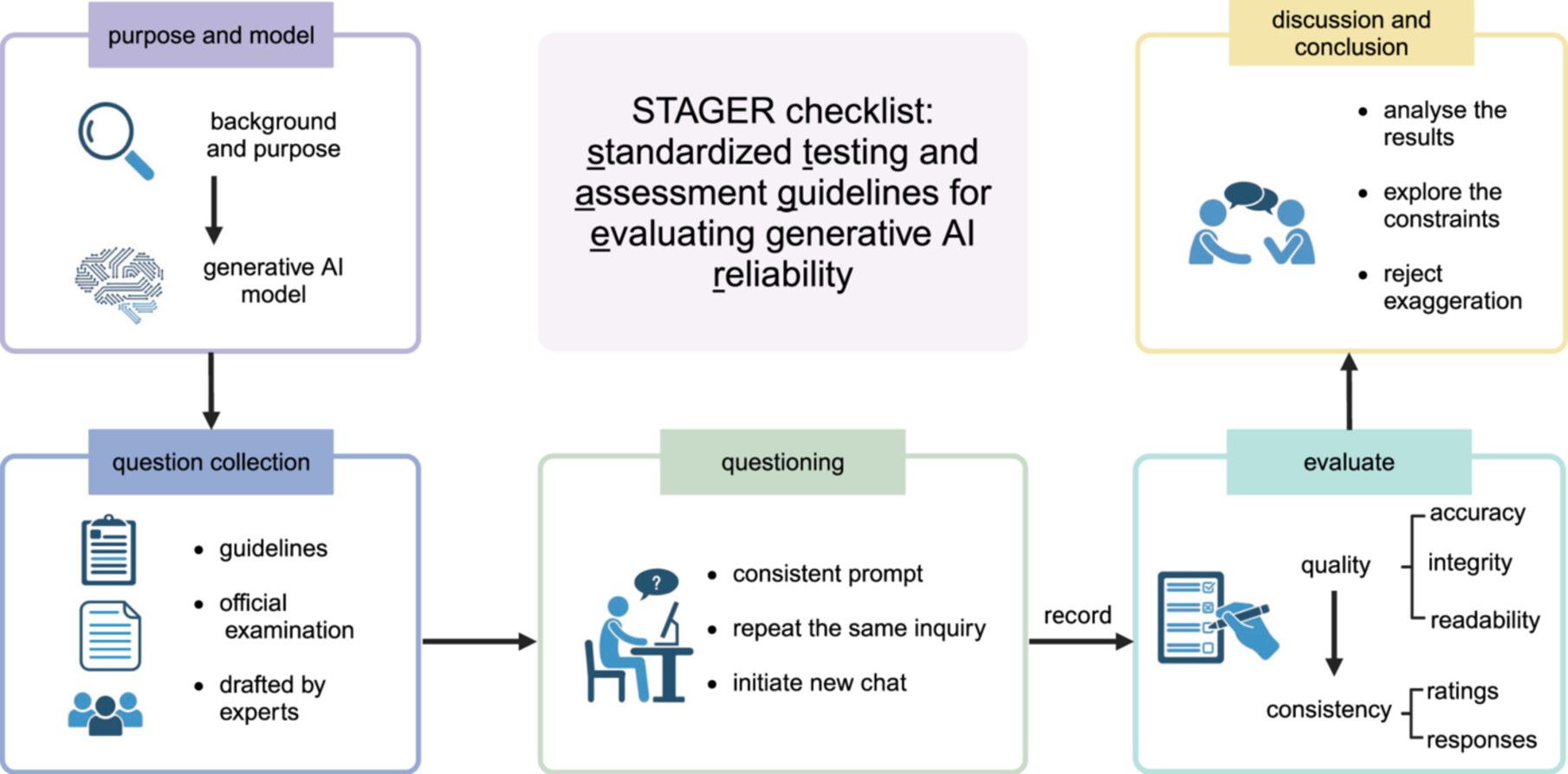
Generative artificial intelligence (AI) holds immense potential for medical applications, but the lack of a comprehensive evaluation framework and methodological deficiencies in existing studies hinder its effective implementation. Standardized assessment guidelines are crucial for ensuring reliable and consistent evaluation of generative AI in healthcare. Our objective is to develop robust, standardized guidelines tailored for evaluating generative AI performance in medical contexts. Through a rigorous literature review utilizing the Web of Sciences, Cochrane Library, PubMed, and Google Scholar, we focused on research testing generative AI capabilities in medicine. Our multidisciplinary team of experts conducted discussion sessions to develop a comprehensive 32-item checklist. This checklist encompasses critical evaluation aspects of generative AI in medical applications, addressing key dimensions such as question collection, querying methodologies, and assessment techniques. The checklist and its broader assessment framework provide a holistic evaluation of AI systems, delineating a clear pathway from question gathering to result assessment. It guides researchers through potential challenges and pitfalls, enhancing research quality and reporting and aiding the evolution of generative AI in medicine and life sciences. Our framework furnishes a standardized, systematic approach for testing generative AI's applicability in medicine. For a concise checklist, please refer to Table S or visit GenAIMed.org.
Comparative multiomics analyses reveal the breed effect on the colonic host–microbe interactions in pig
- 04 July 2024

Dysregulation of the gut microbiota often leads to immune-related disorders, indigestion, or diarrhea. Here, Jiaxing Black (JXB) pig, a local Chinese pig breed known for its great tolerance and digestibility of nutrients, was employed for a metagenomic and transcriptomic integrative analysis to reveal the gut microbiota-genes and gut microbiota-pathway interactions. A total of 452 differentially expressed genes, and 174 phyla were found between the JXB and the Duroc × Landrace × Yorkshire (DLY) pigs. Detailed analysis revealed that the differences in colon gene expression signatures between the JXB and DLY are mainly enriched in metabolic and inflammatory responses, with Lactobacillus and Lachnospiraceae enriched in DLY and JXB, respectively. Notably, Pacebacteria, Streptophyta, and Aerophobetes were found to participate in the PI3K-Akt mediated immune response in both pig breeds; however, they only accelerated the metabolism in the intestines of JXB pigs. Moreover, the host could regulate microbe metabolism and immune response by Ig-like domain-containing protein and ITIH2, PAEP, and TDRD9, respectively. Taken together, our results revealed both common and breed-specific regulations of host genes by gut microbiota in two pig breeds.