The role of the gut microbiome in the regulation of high-altitude adaptation
- 19 January 2026
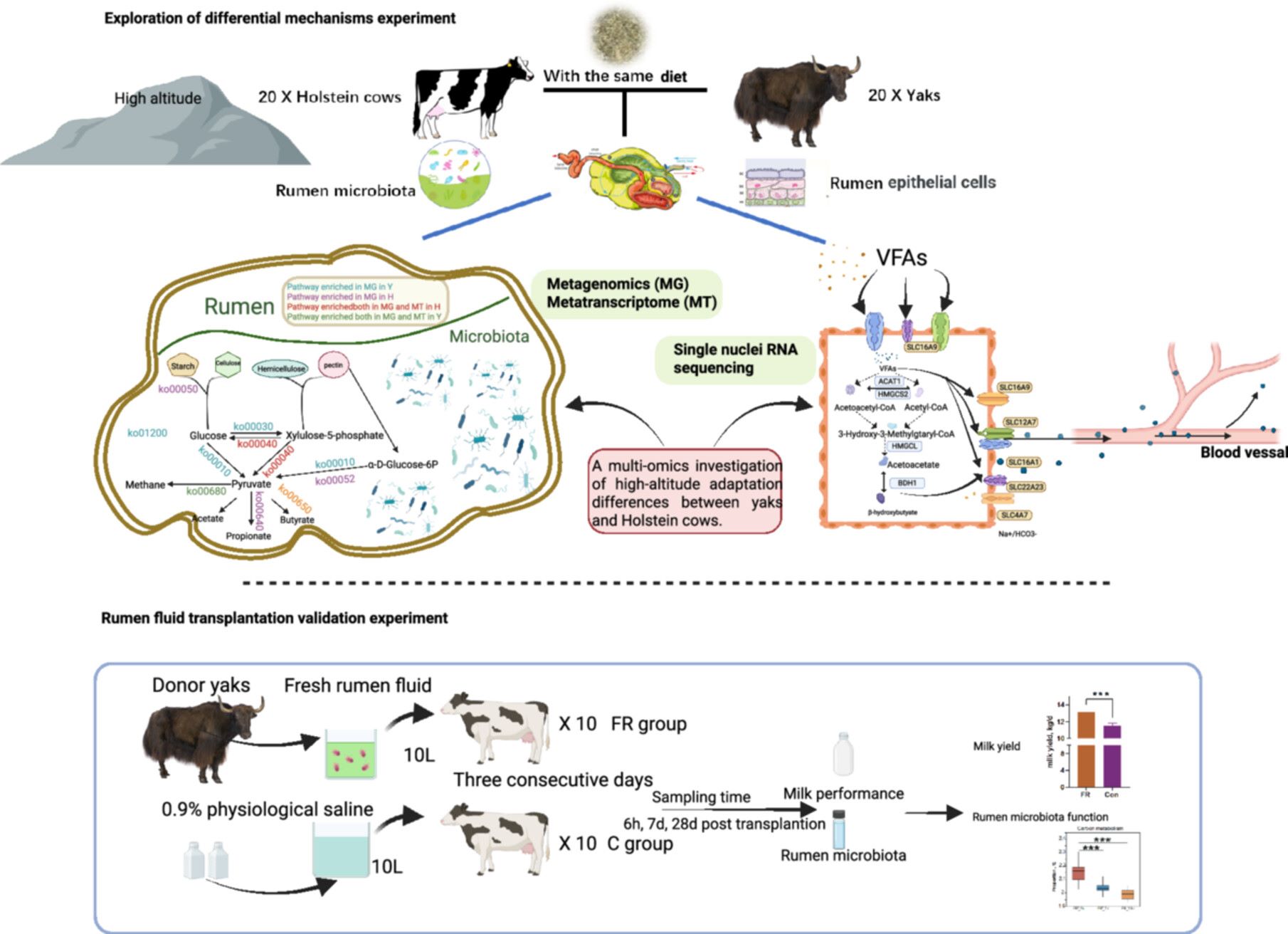
This study is the first to elucidate mechanisms of high-altitude adaptation from the perspective of the rumen ecosystem by using indigenous yaks and Holstein cows that have lived at high altitude since birth as comparative models. Through a systematic comparison of their rumen ecology using multi-omics approaches—including rumen metagenomics, metatranscriptomics, and single-nucleus transcriptomics—we identified distinct adaptive strategies employed by the two species under extreme plateau environments. Specifically, the rumen microbiota of yaks was enriched in central carbon metabolism pathways, whereas the microbiota of Holstein cows was enriched in pathways related to the degradation of specific complex carbohydrates. Concurrently, yak rumen epithelial cells exhibited increased expression of genes associated with ketone body metabolism, along with higher pathway activity scores for ketone body biosynthesis and fatty acid beta-oxidation. Moreover, we introduced rumen fluid transplantation from yaks for the first time and observed that, under the conditions of this study, yak microbiota transplantation increased milk yield in Holstein cows. After transplantation, the carbon metabolism of the rumen microbiota gradually declined, further confirming the key role of the rumen microbiota in regulating host energy supply in the context of yak high-altitude adaptation. Overall, this study deciphers the mechanisms by which gastrointestinal microbiota contributes to high-altitude adaptation in ruminants and provides a new perspective for addressing challenges related to adaptation to high-altitude environments.
Host-driven hepatic conversion of gut microbiota-derived putrescine to spermidine mediates mannose's protective effects against hepatic steatosis in zebrafish
- 20 December 2025
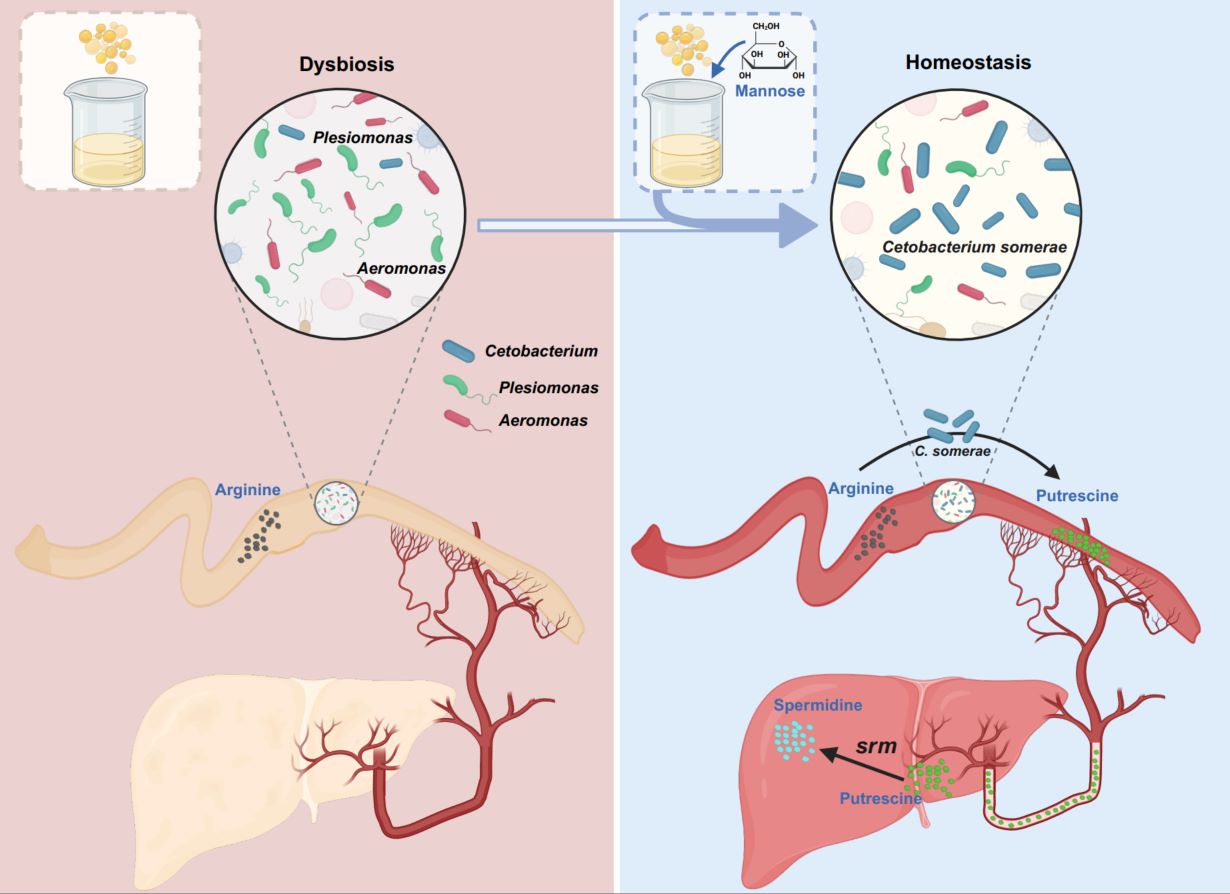
Evidence for liver metabolism of gut-derived microbial compounds into beneficial secondary metabolites has been lacking. Here, we demonstrate that Cetobacterium somerae (C. somerae), enriched by mannose supplementation under high-fat diet conditions, convert arginine into putrescine. The liver subsequently converts this microbial-derived putrescine to spermidine, which functions as an effector molecule to reduce hepatic lipid accumulation. These findings uncover a novel host-microbiota collaborative mechanism in which the arginine-putrescine-spermidine metabolic pathway is completed through inter-kingdom cooperation to ameliorate hepatic steatosis.
Nano-plastics disrupt systemic metabolism by remodeling the bile acid–microbiota axis and driving hepatic–intestinal dysfunction
- 31 December 2025
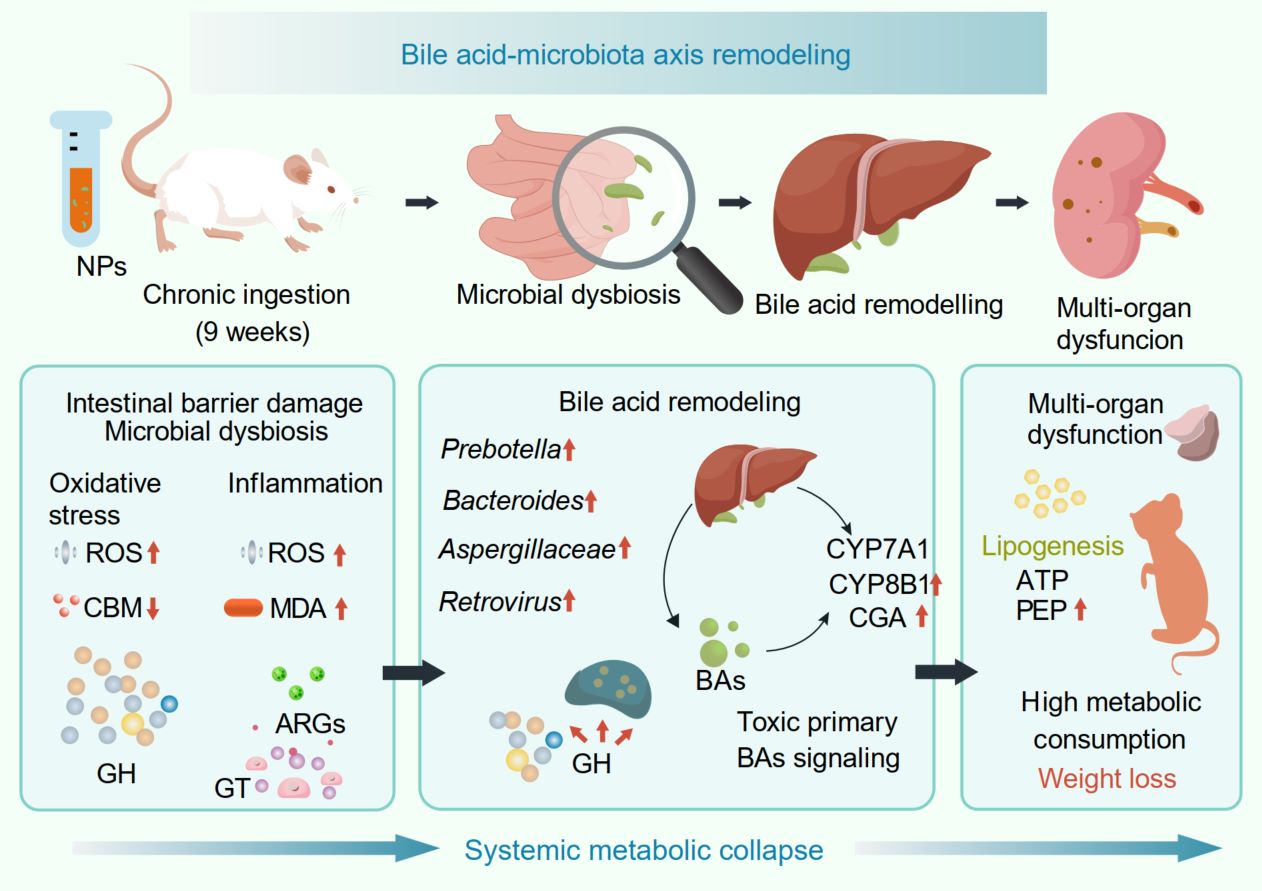
The pervasiveness of microplastic pollution poses a growing health risk, yet its long-term metabolic consequences remain poorly defined. Here, we exposed mice to polyethylene terephthalate nanoparticle (NP) and combined histopathology, biochemistry, metabolomics, and metagenomics to resolve their interactions. NP ingestion induced a severe systemic phenotype characterized by weight loss, organ atrophy, dyslipidemia, and gut barrier collapse. Mechanistically, NPs disrupted bile acid (BA) homeostasis by hyperactivating hepatic synthesis pathways while suppressing microbial 7α-dehydroxylation. This accumulation of cytotoxic BAs drove hepatic lipogenesis and aggravated mucosal inflammation. Crucially, metagenomics uncovered significant gut microbiota dysbiosis, where the enrichment of bile salt hydrolase-encoding taxa and depletion of 7α-dehydroxylating clades reinforced this BA imbalance. Furthermore, the microbiota exhibited functional deterioration, shifting toward glycan degradation with a concurrent loss of antibiotic resistance genes, signaling reduced ecological resilience. These findings identify BA dysregulation and specific microbiota functional losses as primary drivers of NP-induced systemic metabolic collapse.
CAT-BLAST: Engineered bacteria for robust targeting and elimination of cancer-associated fibroblasts
- 05 January 2026
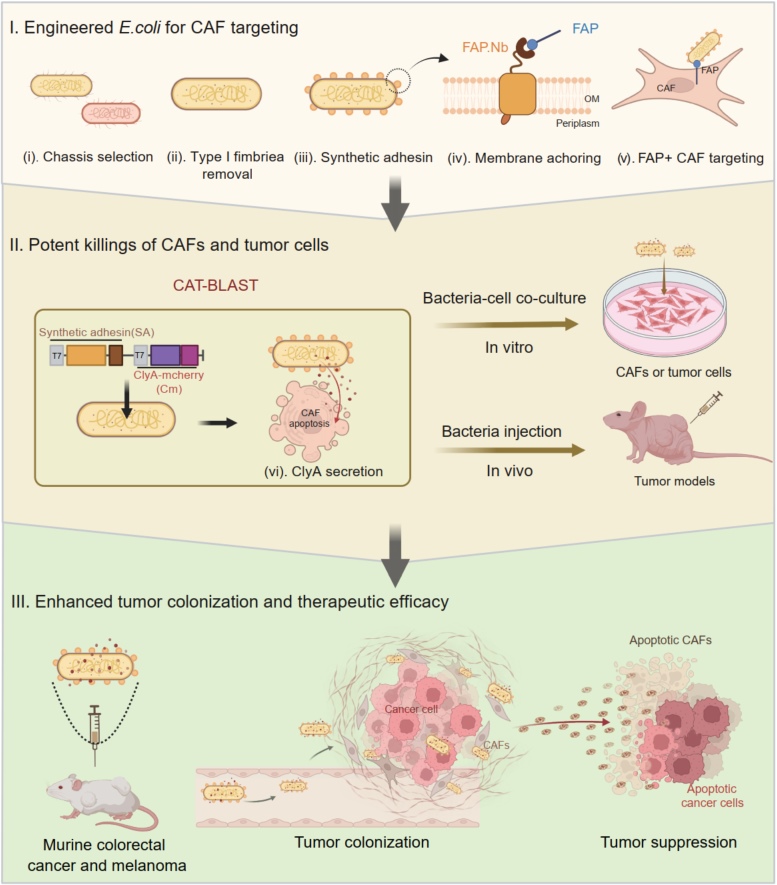
Cancer-associated fibroblasts (CAFs) construct a protective stromal barrier that promotes tumor growth and resistance to therapy. To dismantle this, we developed CAT-BLAST, an engineered bacterial platform designed to potently eliminate these cells. We engineered a safe, high-expression E. coli chassis, arming it with a FAP-specific synthetic adhesin for precise CAF targeting and the ability to secrete the ClyA cytotoxin to induce apoptosis in both CAFs and adjacent tumor cells. This platform demonstrated robust, FAP-specific targeting across diverse human tumor xenograft models and achieved significant tumor suppression in both murine colorectal cancer and melanoma.
A vast stem-progenitor cell pool, richly vascular system, and hybrid ossification drive the daily centimeter-scale elongation of bony antlers
- 08 December 2025
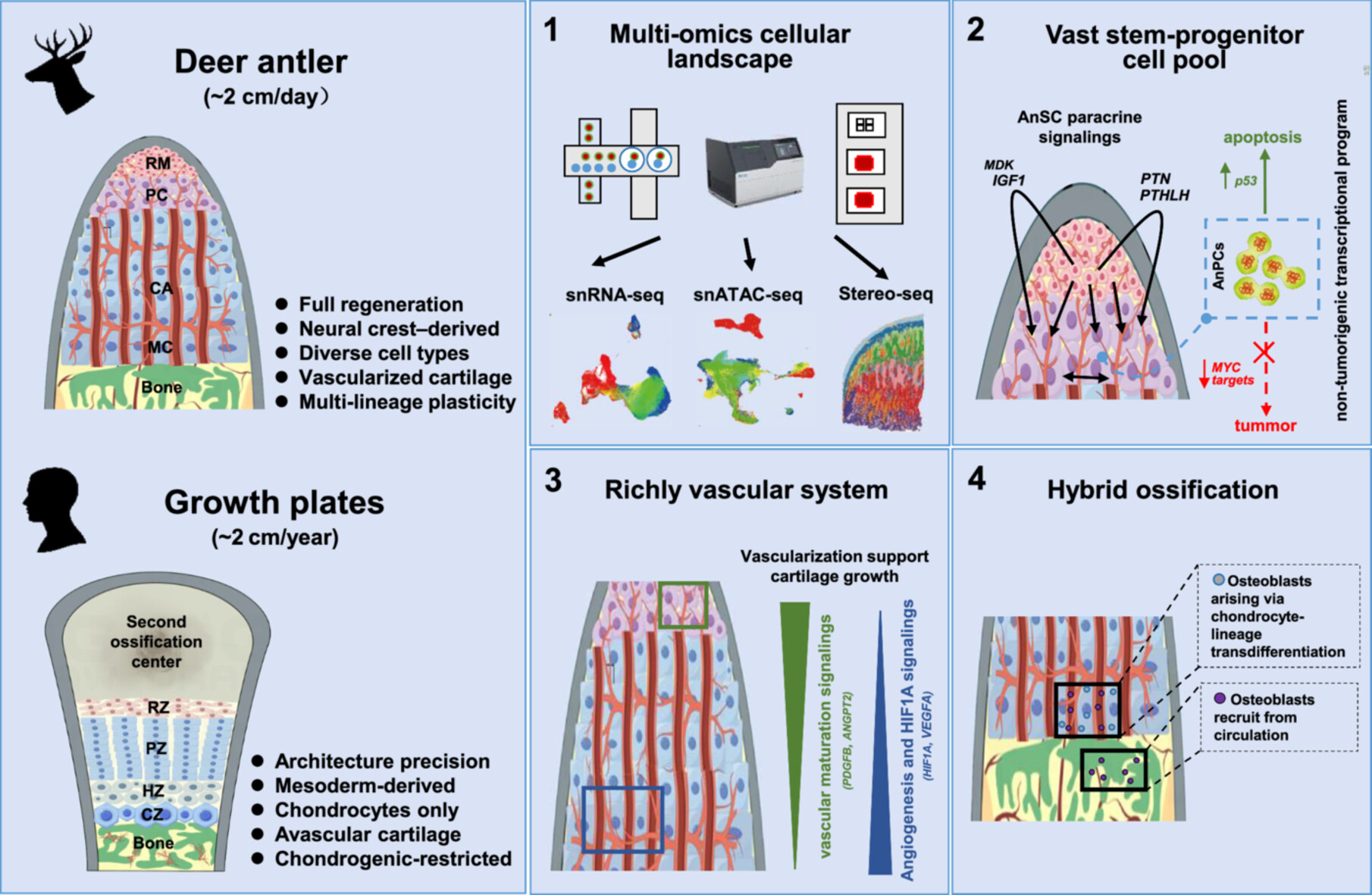
Bone growth and regeneration remain major clinical challenges. Deer antlers, the fastest-growing mammalian bone, regenerate via endochondral ossification and elongate up to 2 cm per day, far surpassing the ~2 cm annual growth of human growth plates. Here, we systematically mapped the cellular landscape of the antler growth center (AGC) using single-nucleus RNA sequencing, chromatin accessibility profiling, and spatial transcriptomics. The AGC harbors a large stem-progenitor pool that drives rapid elongation through vigorous proliferation supported by paracrine signaling. These proliferative cells exhibit a transcriptional program with intrinsically low tumorigenic potential, associated with apoptotic regulation. The AGC also establishes a vascularized niche that supports robust angiogenesis, sustains accelerated cartilage growth, and enables efficient recruitment of osteogenic cells. Notably, antlers employ a hybrid ossification strategy, combining endochondral ossification with direct hypertrophic chondrocyte-to-osteoblast transdifferentiation, likely via PHEX⁺ intermediates. Collectively, these findings refine fundamental concepts of endochondral ossification and offer insights for regenerative bone therapies.
Single-cell spatial transcriptomics reveals potential molecular mechanisms of Abelmoschus manihot (L.) medic in treating diabetic kidney disease
- 08 December 2025
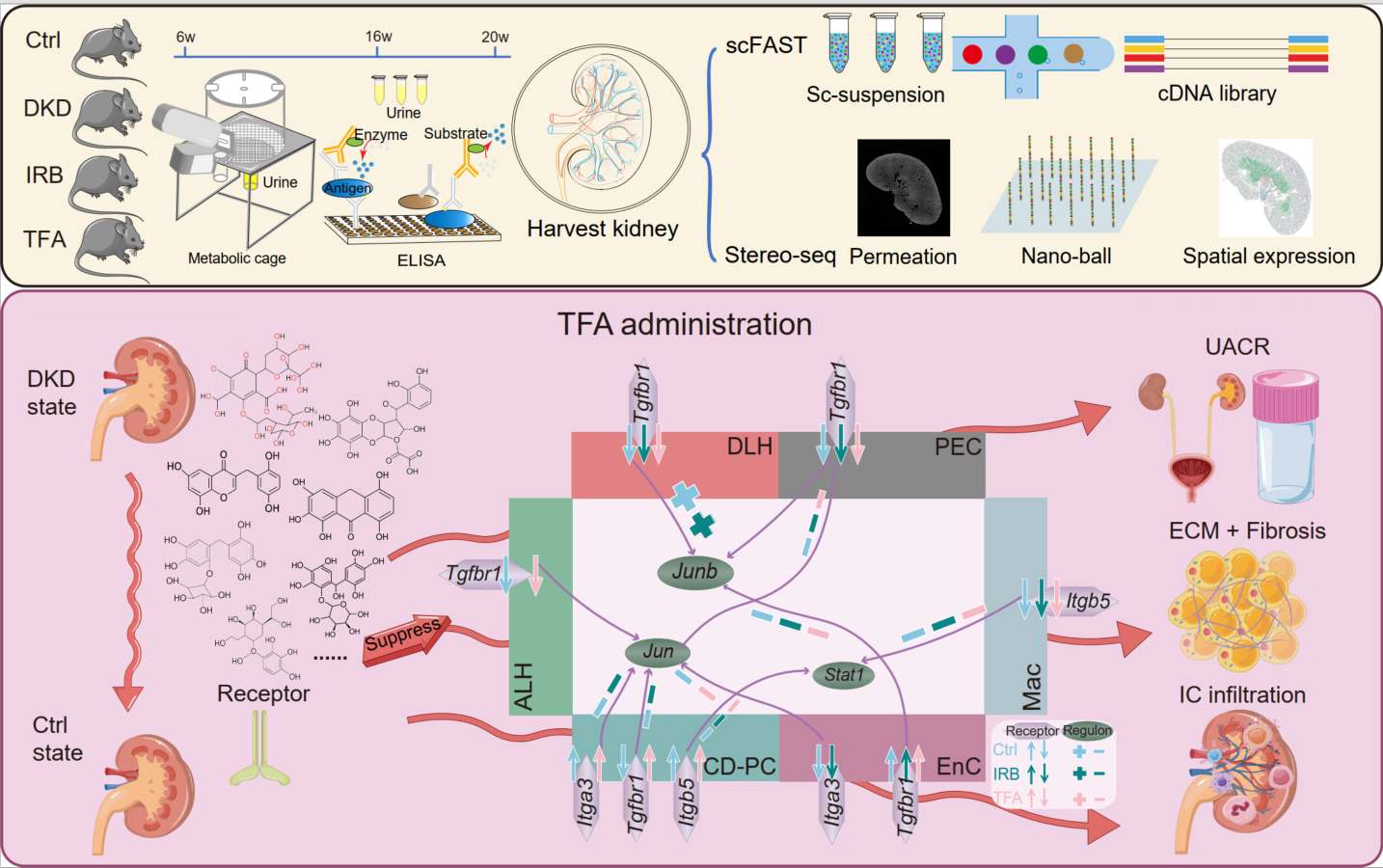
A clinical study reported that Abelmoschus manihot (L.) Medic (A. manihot), in the form of Huangkui capsule (HKC), combined with irbesartan (IRB) is an effective therapy for patients with diabetic kidney disease (DKD). The bioactive components of HKC are total flavones extracted from A. manihot (TFA). To explore the pharmaceutical molecular mechanisms underlying the efficacy of A. manihot in the treatment of DKD, we have combined SpaTial Enhanced REsolution Omics-sequencing (at 0.25 μm resolution) with single-cell full-length RNA sequencing. We employed the db/db mouse model of type 2 diabetes and DKD. These experimental methods generated the first single-cell resolution pharmacopathological spatial atlas in kidneys of db/db mice that were treated with TFA or IRB. TFA exhibited therapeutic effects on DKD comparable to those of TFA combined with IRB. Following genome-wide gene screening and molecular docking simulation, we have identified the key renal receptors (Itga3, Itga5, Tgfbr1, etc.) and regulators (Jun, Junb, Stat1, etc.) underlying the therapeutic action of TFA in DKD. This study provides novel insights into the pharmaceutical mechanisms of A. manihot in the treatment of DKD.
Genome-wide CRISPR screen reveals an uncharacterized spliceosome regulator as new candidate immunotherapy target
- 28 November 2025
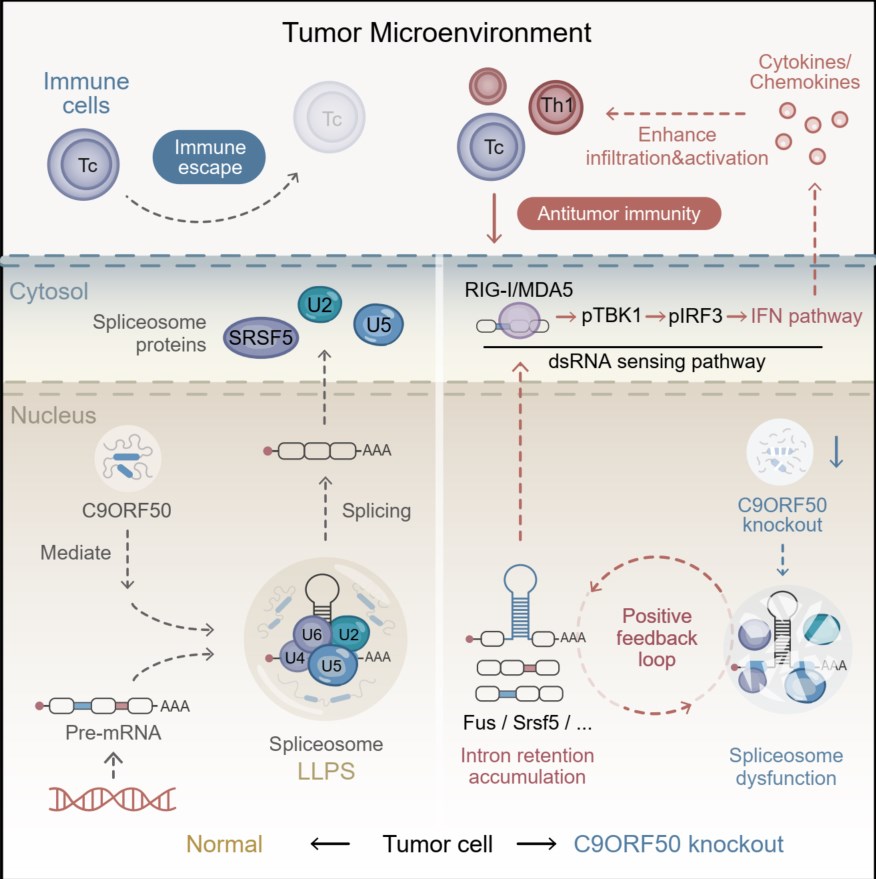
This study identifies the uncharacterized gene C9ORF50 as a novel regulator of immune evasion. Functioning as an intrinsically disordered protein, C9ORF50 drives liquid-liquid phase separation to facilitate spliceosome assembly and maintain RNA splicing fidelity. Genetic ablation or therapeutic inhibition of C9ORF50 disrupts this process, leading to widespread intron retention in pre-mRNAs of core spliceosome components. These unprocessed introns form immunogenic double-stranded RNA (dsRNA) that accumulates in the cytoplasm. This dsRNA is sensed by innate immune receptors (RIG-I/MDA5), activating the TBK1-IRF3 axis and subsequent type I interferon signaling. Consequently, this enhances the production of T-cell chemoattractants (CXCL9/10/11), promoting robust CD8⁺ and CD4⁺ T cell infiltration into tumors. Together, these findings reveal that C9ORF50 inhibition effectively converts immune-suppressive tumors into T cell-inflamed lesions, thereby enhancing immunogenicity and suppressing tumor growth.
Soil-borne legacy facilitates the dissemination of antibiotic resistance genes in soil–plant continua
- 23 November 2025
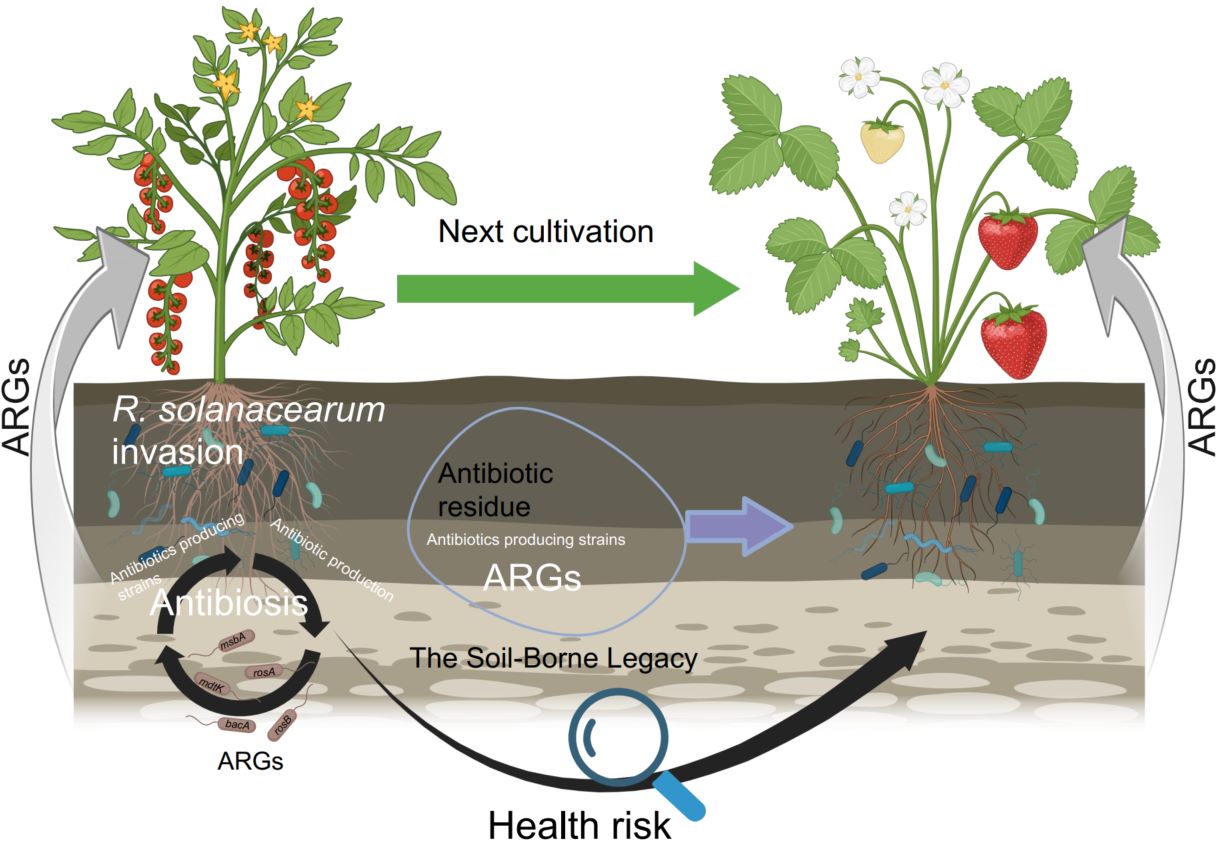
Antimicrobial resistance (AMR) disseminates throughout the soil–plant continuum via complex microbial interactions. Plants shape root- and leaf-associated microbiomes that sustain plant health; however, soil-borne legacies—enriched with antibiotic-producing microbes and resistance genes—govern AMR dynamics across agroecosystems. Using 16S rRNA gene sequencing, shotgun metagenomics, and high-throughput quantitative PCR, we profiled antibiotic resistance genes (ARGs), mobile genetic elements, and virulence factor genes across bulk soil, rhizosphere, phyllosphere, and root endosphere within soil–tomato and soil–strawberry continua. Recurrent bacterial wilt amplified the resistome, particularly polypeptide resistance genes, thereby establishing the rhizosphere as a major hotspot of ARG accumulation. Multidrug-resistant Ralstonia solanacearum (R. solanacearum) strains acted as major ARG reservoirs, harboring resistance determinants on both chromosomes and megaplasmids. Collectively, these findings demonstrate that pathogen-driven restructuring of the plant microbiome accelerates ARG dissemination, establishing soil-borne diseases as critical amplifiers of AMR across agricultural ecosystems.
PhyloSuite v2: The development of an all-in-one, efficient and visualization-oriented suite for molecular dating analysis and other advanced features
- 25 November 2025
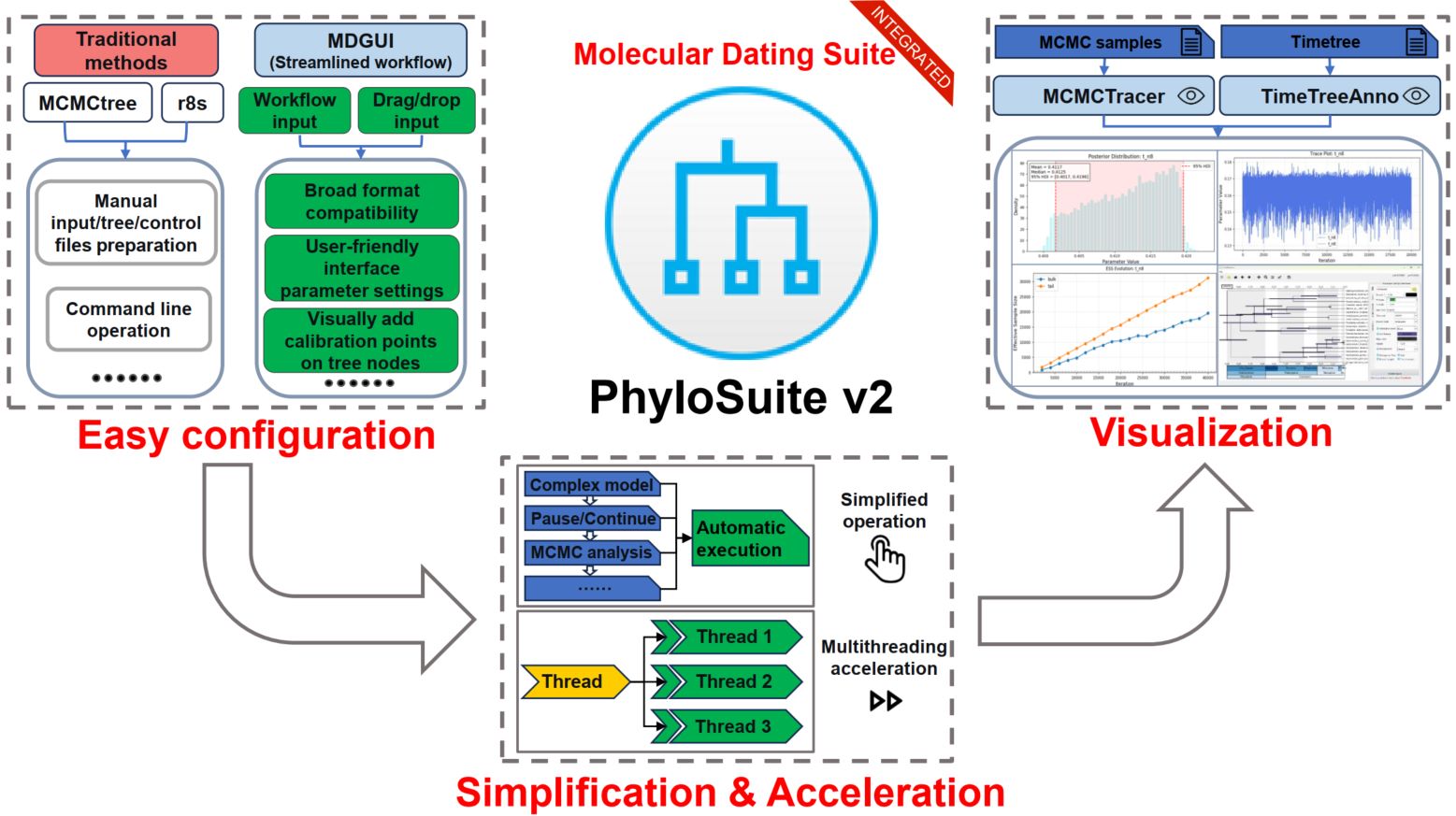
MCMCtree and r8s are among the most popular molecular dating tools in the current genomic era, but their utility is hampered by steep learning curves, particularly concerning input file formatting, the complexity of fossil calibration setup, tree visualization, and model selection. To enhance their usability and improve research efficiency, we developed three new tools: MDGUI (for molecular dating analysis), TimeTreeAnno (for timetree visualization), and MCMCTracer (for convergence assessment). We integrated these into the PhyloSuite v2 platform, along with MCMCtree and r8s plugins, to create a comprehensive molecular dating suite. Compared to existing solutions that we benchmarked, our toolkit offers a more intuitive interface and streamlined workflow, featuring visual calibration point configuration, support for multiple alignment formats, automated model selection and implementation for downstream analyses, one-click pause/resume functionality, multithreading acceleration, and on-demand MCMC convergence assessment and plotting. Furthermore, PhyloSuite v2 introduces other advanced features, including gene duplicate resolution during the extraction step, significantly accelerated data handling capabilities (specifically, format conversion and concatenation), deeper integration of the latest IQ-TREE models and functions, and further streamlining of the entire phylogenetic analysis workflow. The update also includes adaptation to high-resolution screens and numerous bug fixes. The source code for the new version of PhyloSuite is available at https://github.com/dongzhang0725/PhyloSuite.
The interplay between tissue-resident microbiome and host proteins by integrated multi-omics during progression of colorectal adenoma to carcinoma
- 04 November 2025
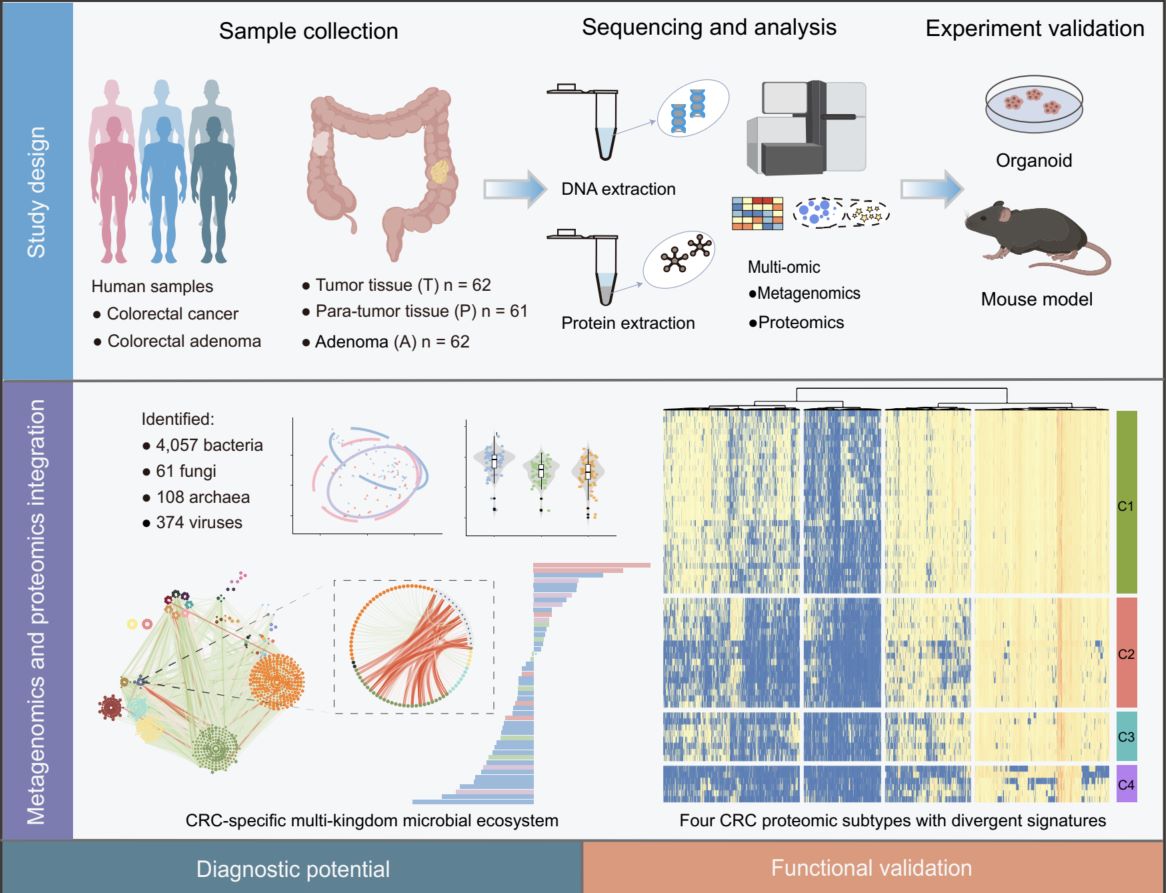
This study integrated metagenomic and proteomic profiling of 185 colorectal tissue samples—spanning adenoma (A), tumor (T), and para-tumor (P)—to characterize multi-kingdom microbiome and host protein dynamics in colorectal cancer (CRC). In total, 4057 bacterial, 61 fungal, 108 archaeal, and 374 viral species were identified, revealing CRC-specific microbial dysbiosis. Proteomic analysis defined four molecular subtypes (C1–C4) with distinct clinical outcomes. This study further developed a diagnostic model based on 14 microbial and 8 protein markers, which robustly distinguished adenoma from tumor (achieved an area under the receiver operating characteristic curve (AUROC) = 0.962) and early from advanced stages (AUROC = 0.926), with validation across multiple independent cohorts. Functional assays in patient-derived organoids and murine allografts confirmed that enterotoxigenic Bacteroides fragilis and Fusobacterium nucleatum promote tumor growth through Wnt/β-catenin and NF-κB pathway activation. These findings highlight dynamic host–microbiome interactions in CRC progression and provide novel biomarkers and therapeutic targets.
Whole microbiota transplantation restores gut homeostasis throughout the gastrointestinal tract
- 11 November 2025
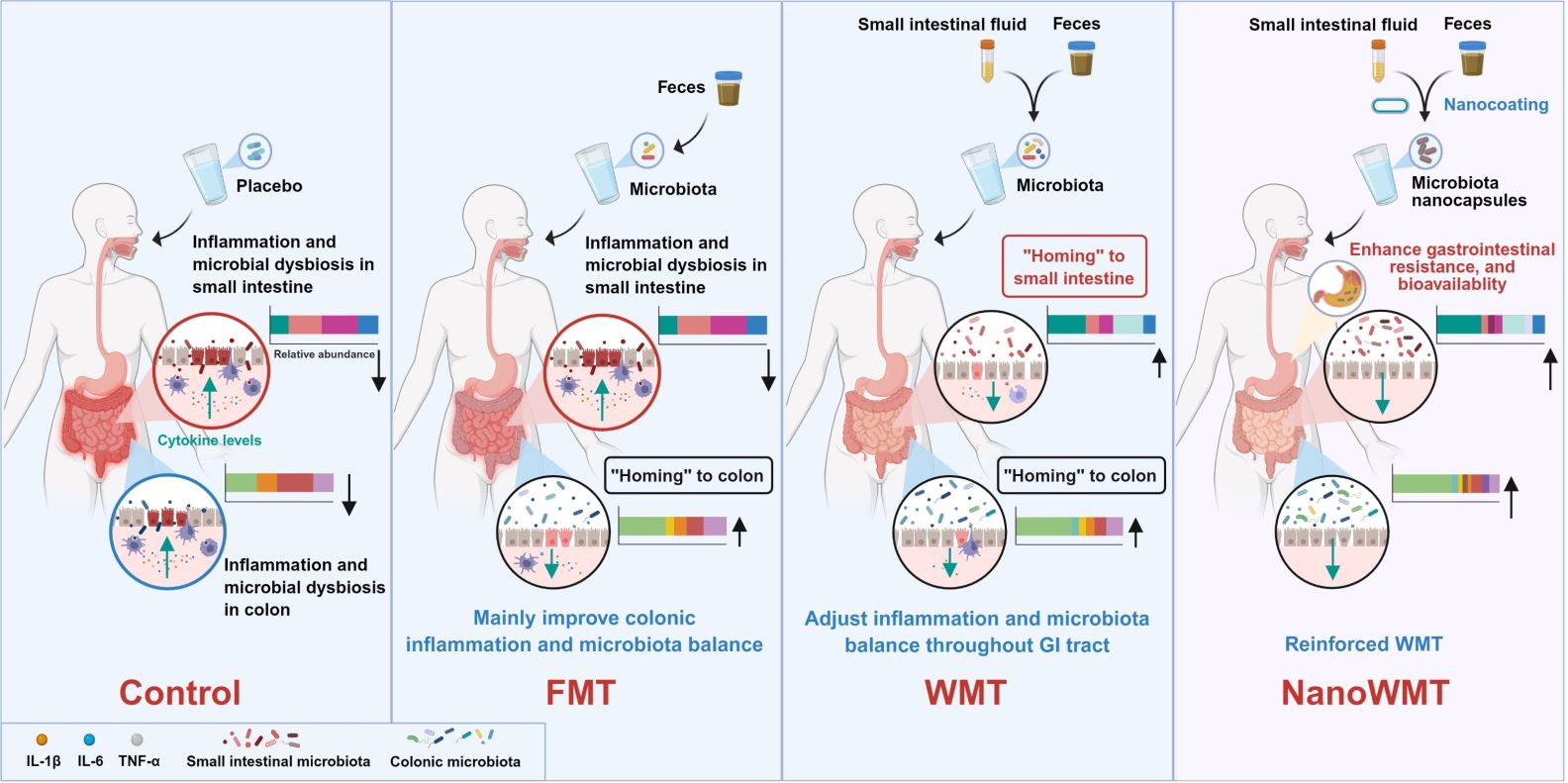
This study introduces whole microbiota transplantation (WMT), a synergistic therapeutic approach that concurrently transplants small intestinal and fecal microbiota. In germ-free mice, WMT outperforms conventional fecal microbiota transplantation (FMT) in restoring gut microbiota diversity and abundance. Moreover, in a chemotherapy-induced intestinal mucositis model, WMT alleviates intestinal inflammation and reverses microbiota dysbiosis. Encapsulation in layer-by-layer self-assembled nanocapsules further boosts microbial survival and colonization, amplifying WMT's anti-inflammatory effects and microbiota restoration in a mouse model of pan-intestinal infection. Overall, WMT represents a precise strategy for reshaping microbial homeostasis across the entire gastrointestinal tract, with therapeutic promise for inflammatory bowel diseases and small-intestinal disorders.
High-throughput generic single-entity sequencing using droplet microfluidics
- 30 October 2025
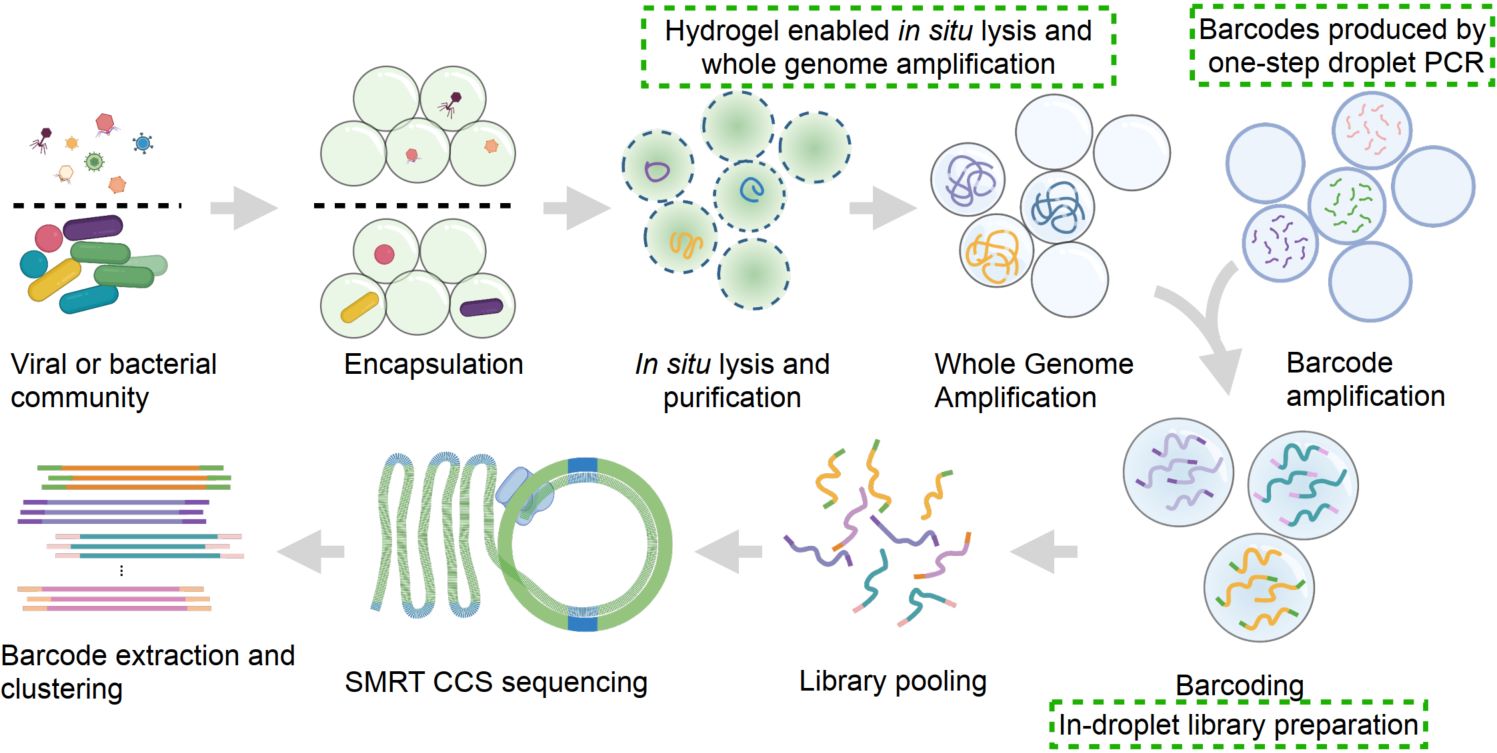
We present Generic Single Entity Sequencing (GSE-Seq), a droplet-based workflow that generates monoclonal barcodes by one-step PCR, performs dissolvable hydrogel-enabled reactions for genome processing, and attaches barcodes during in-droplet library preparation. Barcoded fragments are pooled and sequenced with PacBio long-read sequencing. Barcodes are extracted and clustered to recover single-entity genomes from viruses and bacteria, enabling high-throughput single-entity sequencing with long-read and accurate genome reconstruction.
Homoharringtonine suppresses acute myeloid leukemia progression by orchestrating EWSR1 phase separation in an m6A-YTHDF2-dependent mechanism
- 30 October 2025
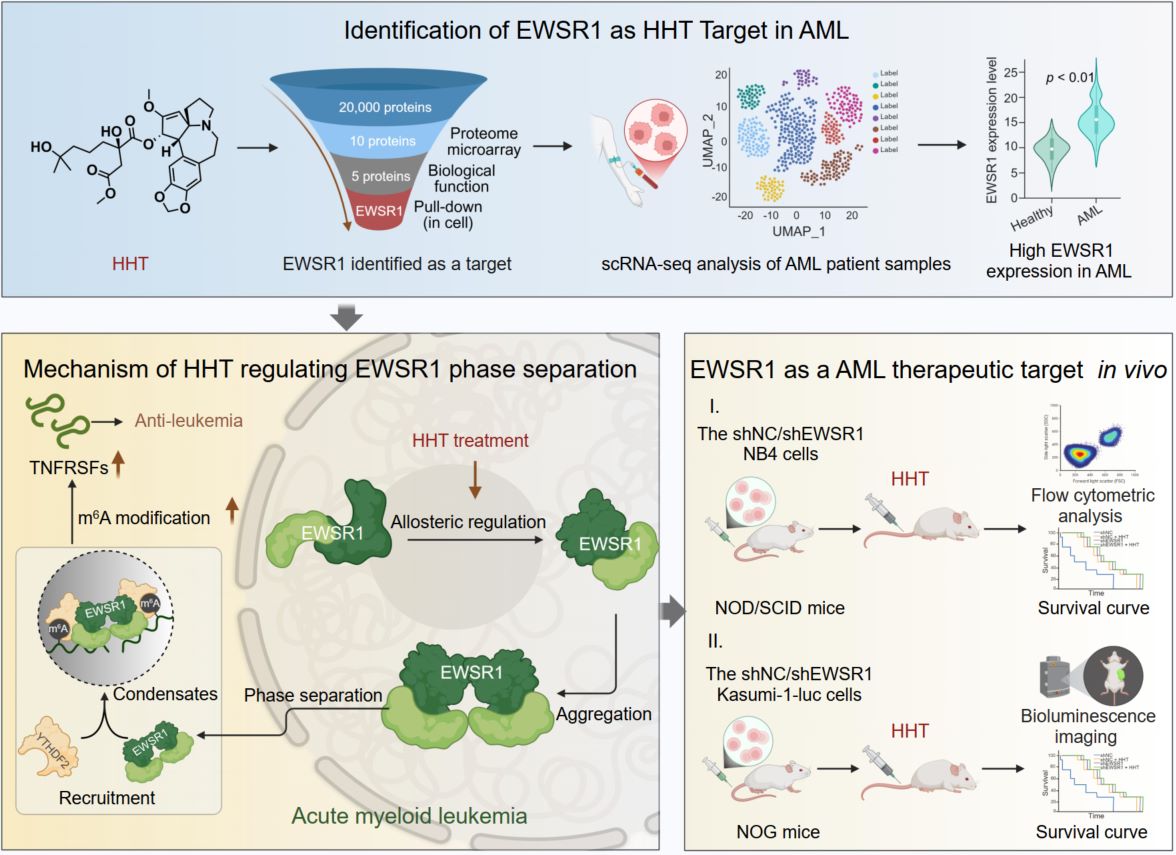
Homoharringtonine (HHT), a clinically approved anti-leukemic drug, directly binds the RNA-binding protein EWSR1. HHT induces an allosteric conformational switch in the RNA recognition motif of EWSR1, driving self-association and liquid–liquid phase separation (LLPS). The resulting condensates act as regulatory hubs that sequester the m6A reader YTHDF2, thereby preventing recognition and decay of m6A-modified transcripts. Stabilization of apoptosis-associated RNAs such as TNFRSF1B and HMOX1 disrupts leukemic cell homeostasis and suppresses AML proliferation. These findings identify EWSR1 as the direct effector of HHT and a predictive biomarker for therapeutic responsiveness in AML.
Nationwide profiling of vaginal microbiota in Chinese women reveals age-dependent shifts and predictive biomarkers for reproductive health
- 23 October 2025
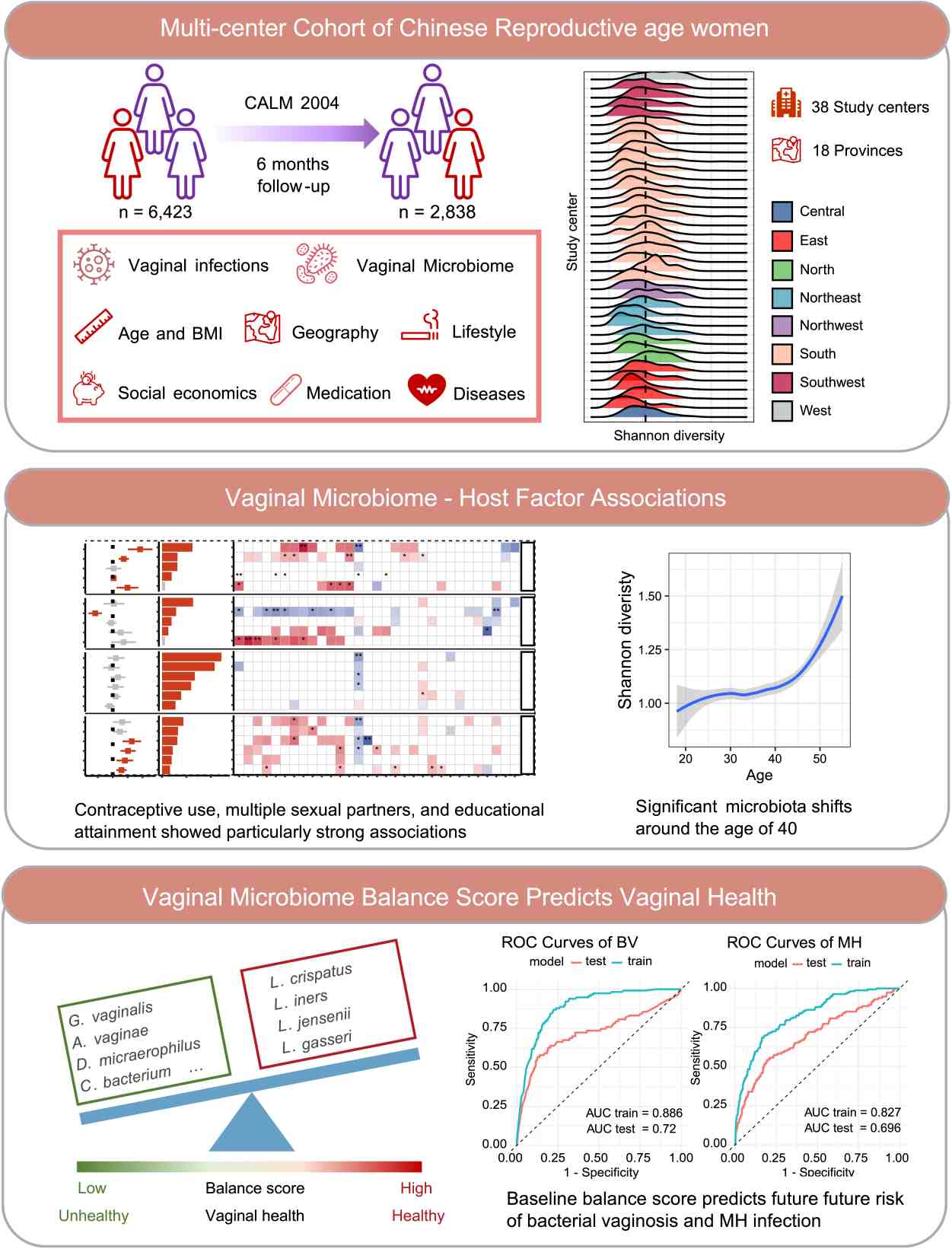
The vaginal microbiome is central to reproductive health, yet large-scale studies in East Asian populations remain scarce. Here, we characterized the vaginal microbiota of 6423 Chinese women of reproductive age across 18 provinces and assessed associations with 33 host factors. We observed a striking compositional transition around age 40, marked by declining Lactobacillus crispatus and enrichment of dysbiosis-associated taxa including Gardnerella vaginalis, independent of lifestyle or sociodemographic influences. Sexual behavior, contraceptive use, and educational attainment emerged as key determinants of community structure, differentially shaping Lactobacillus crispatus and Lactobacillus iners. Despite these associations, host factors explained less than 2% of overall variation, highlighting the resilience and individuality of the vaginal microbiome. To quantify vaginal health, we derived a microbiome balance score, validated it in external cohorts, and demonstrated its predictive power for incident bacterial vaginosis and sexually transmitted infections. Our findings establish a national-scale reference for the vaginal microbiome in Chinese women, reveal a midlife inflection point in microbial composition, and introduce a clinically actionable metric for risk stratification. These insights advance mechanistic understanding of host–microbiome interactions and inform strategies for precision interventions to preserve vaginal health.
iMeta Conference 2025: Creating high-impact international journals
- 15 October 2025

The iMeta Conference 2025, part of the iMeta Conference series, themed “Creating High-Impact International Journals,” held at the Huangjiahu Campus of Hubei University of Chinese Medicine from August 23rd to 25th, 2025, and focused on frontier topics such as microbiology, medicine, traditional Chinese medicine, botany, and research career development. The event aimed to support the development of researchers and strengthen the impact of academic journals. Through invited reports, thematic seminars, and poster presentations, the conference highlighted hot topics including multi-omics technologies, microbe-host interactions, AI-assisted research, live biotherapeutic products, and the modernization of traditional Chinese medicine. The event demonstrated the innovative momentum of interdisciplinary integration and technological convergence, providing an international platform for academic exchange and laying a foundation for building an innovative scientific research ecosystem and enhancing the global influence of Chinese academic journals.
A comprehensive benchmarking for spatially resolved transcriptomics clustering methods across variable technologies, organs, and replicates
- 09 October 2025
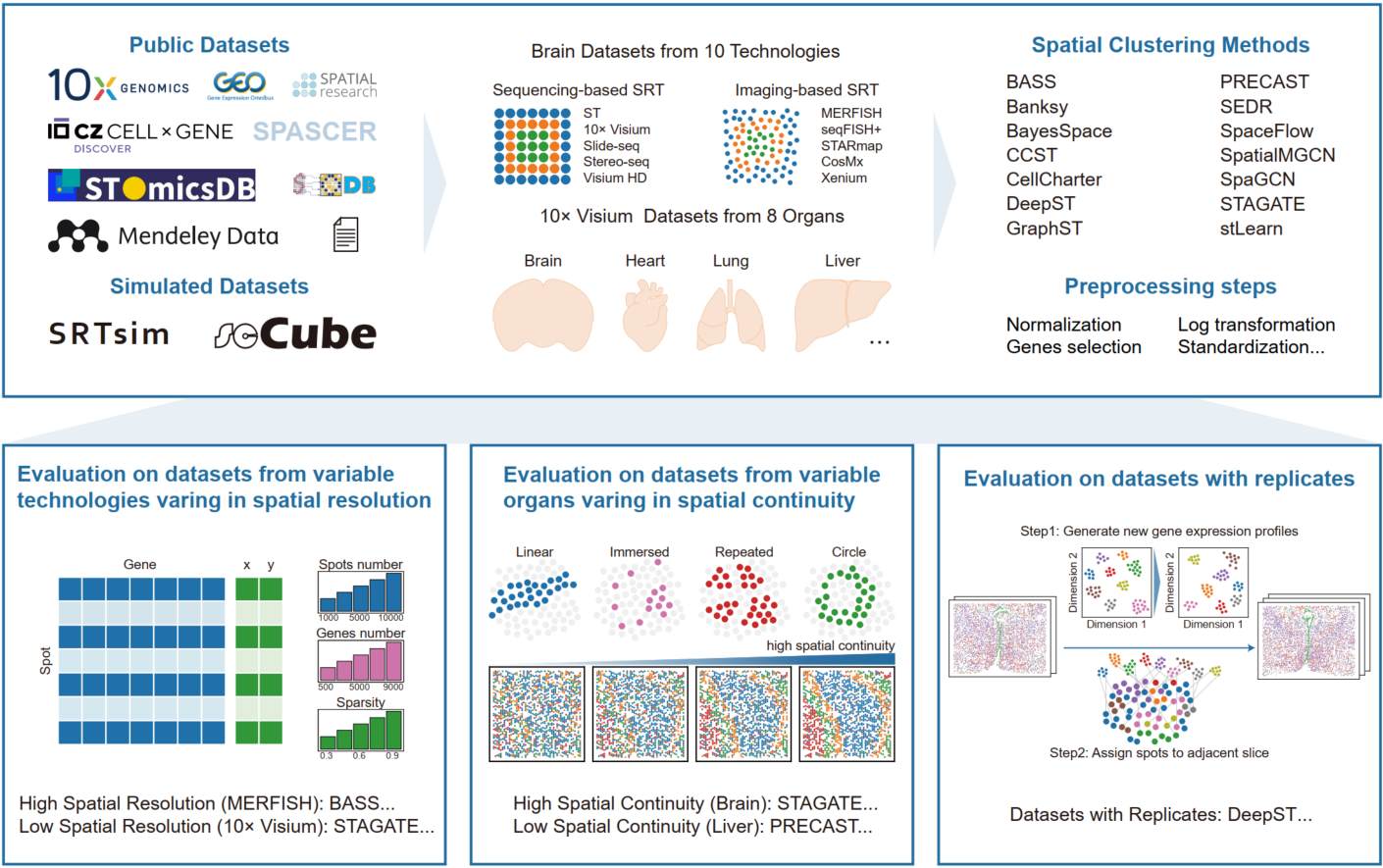
Spatial clustering is a critical step in spatially resolved transcriptomics. Here, we conducted a comprehensive benchmarking analysis of 14 spatial clustering methods using ~600 datasets, including both real-world and simulated data spanning ten technologies and eight organs. We provide practical recommendations for method selection across diverse technologies, organs, and biological replicates. Performance differences are largely explained by data characteristics, spatial patterns, and preprocessing strategies, and we establish an optimized preprocessing pipeline to enhance clustering robustness.
Metagenomics and digital cell modeling facilitate targeted high-throughput sorting of anaerobic hydrogen-producing microorganisms
- 21 September 2025
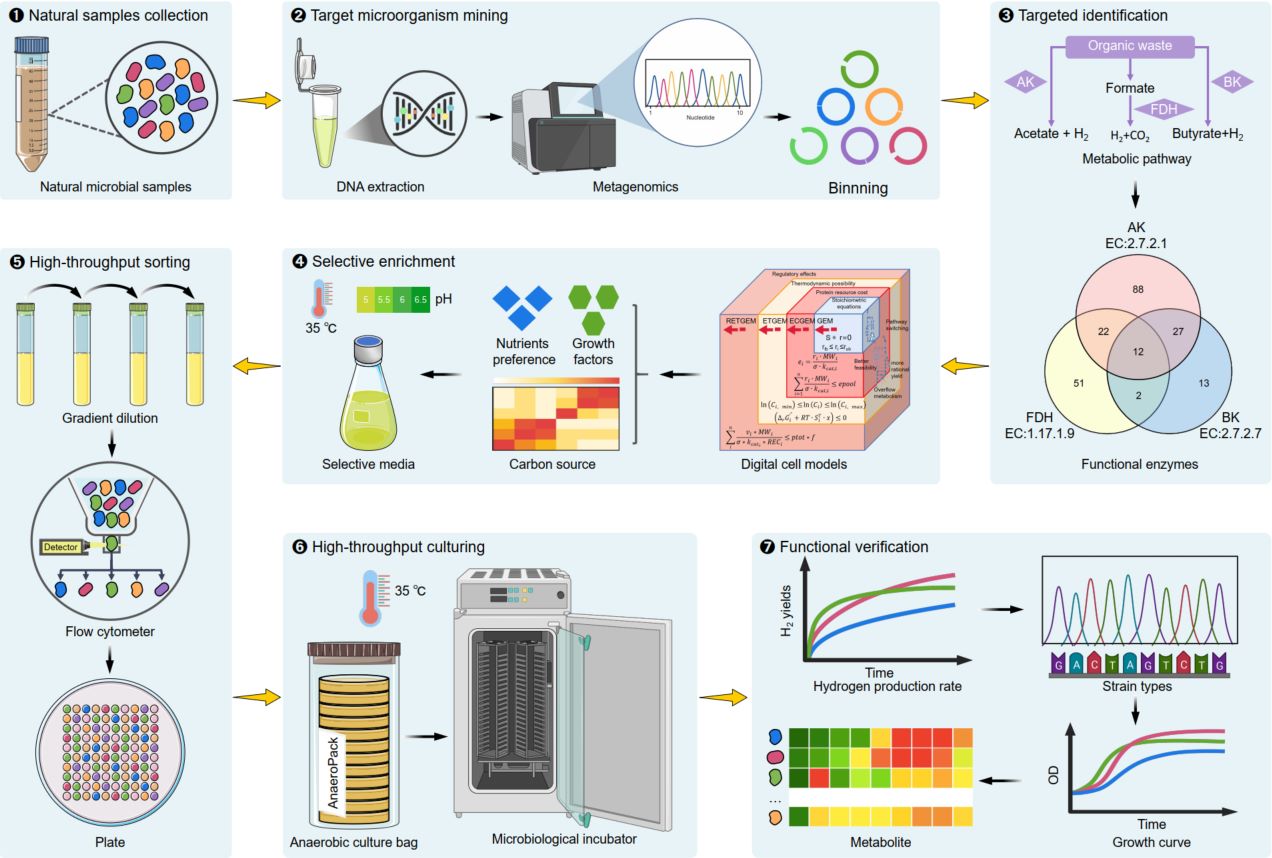
This study proposes a novel strategy that prioritizes functional recognition, followed by targeted high-throughput sorting, to enable the comprehensive, rapid, and efficient acquisition of target microorganisms. Using metagenomic sequencing and binning analysis, we identified 215 potential anaerobic hydrogen-producing strains from 12 large-scale biogas samples. Digital cell models were subsequently constructed from metagenome-assembled genomes, which guided the design of 14 selective culture media for enriching these hydrogen-producing bacteria. Flow cytometry-based high-throughput sorting successfully isolated 81 potential anaerobic hydrogen-producing strains, achieving a target acquisition rate above 37% and a survival rate exceeding 70%. This method holds broad potential for the discovery and sorting of functional microorganisms across diverse environments and may ultimately facilitate the development of synthetic microbiomes for industrial applications.
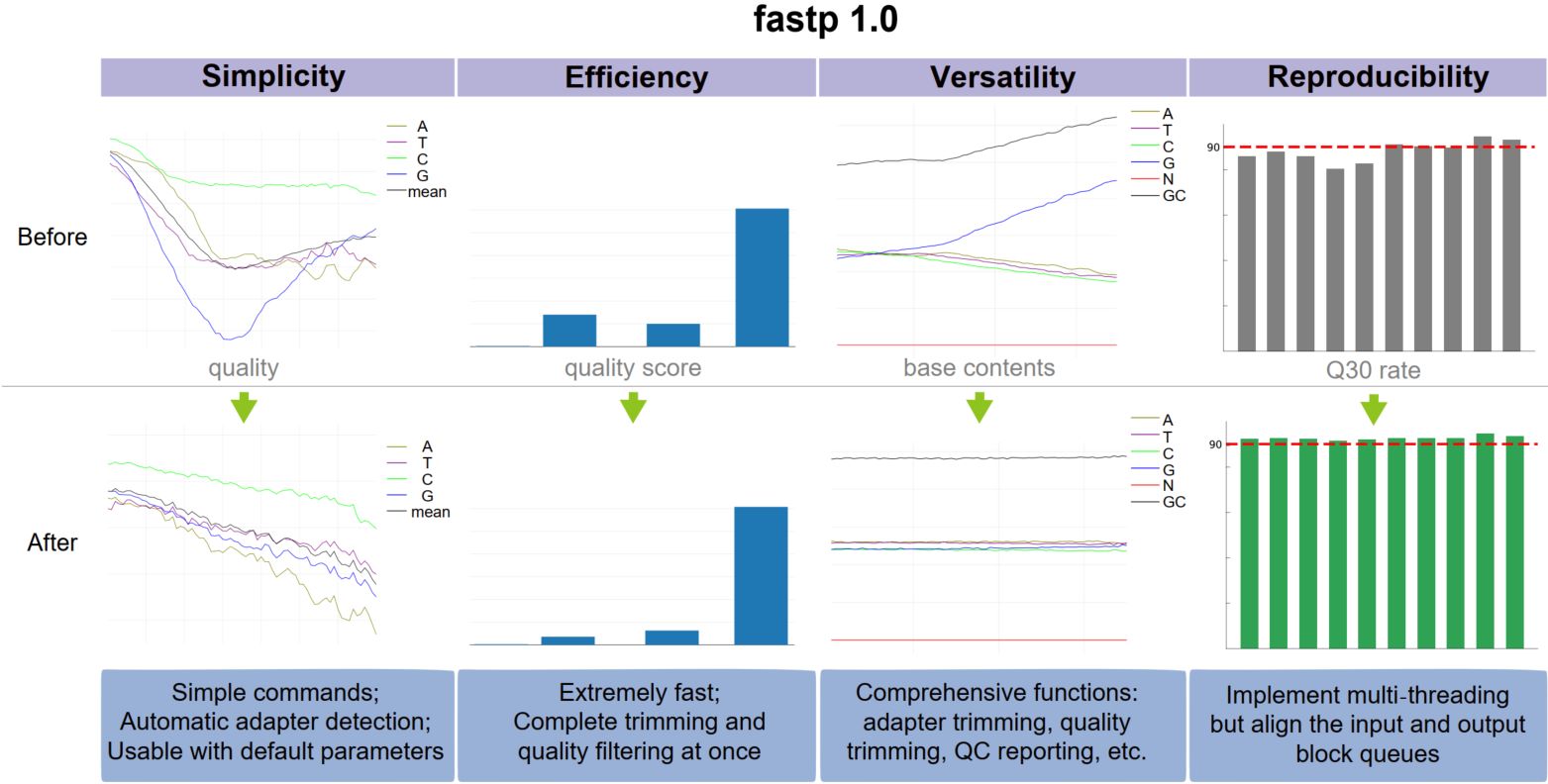
fastp 1.0: An ultra-fast all-round tool for FASTQ data quality control and preprocessing
- 09 September 2025
EasyMetagenome: A user-friendly and flexible pipeline for shotgun metagenomic analysis in microbiome research
- 14 February 2025
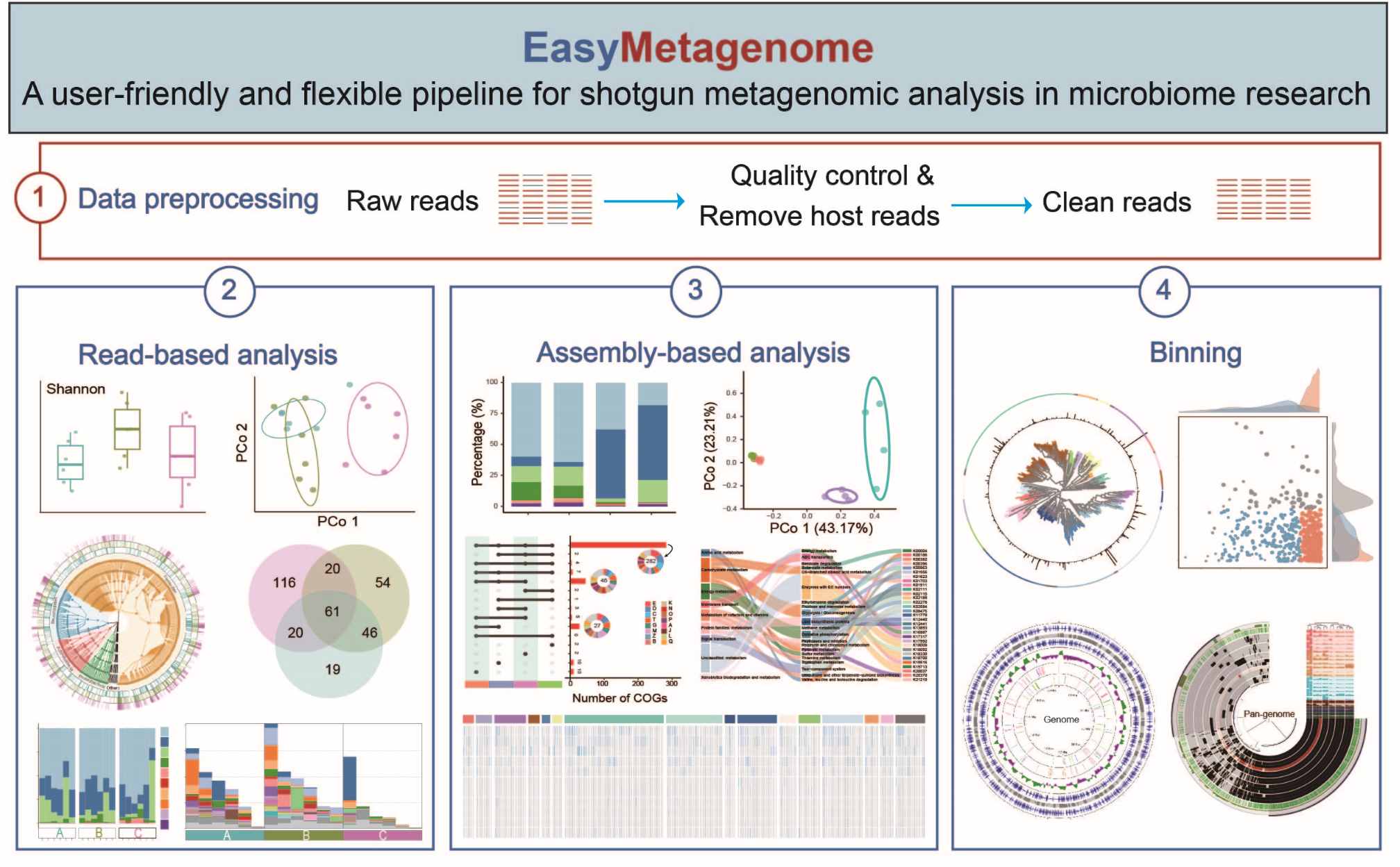
EasyMetagenome is a user-friendly shotgun metagenomics pipeline designed for comprehensive microbiome analysis, supporting quality control, host removal, read-based, assembly-based, binning, genome and pan-genome analysis. It offers customizable settings, data visualizations, and parameter explanations. The pipeline is freely available at https://github.com/YongxinLiu/EasyMetagenome.
Gut–X axis
- 26 February 2025
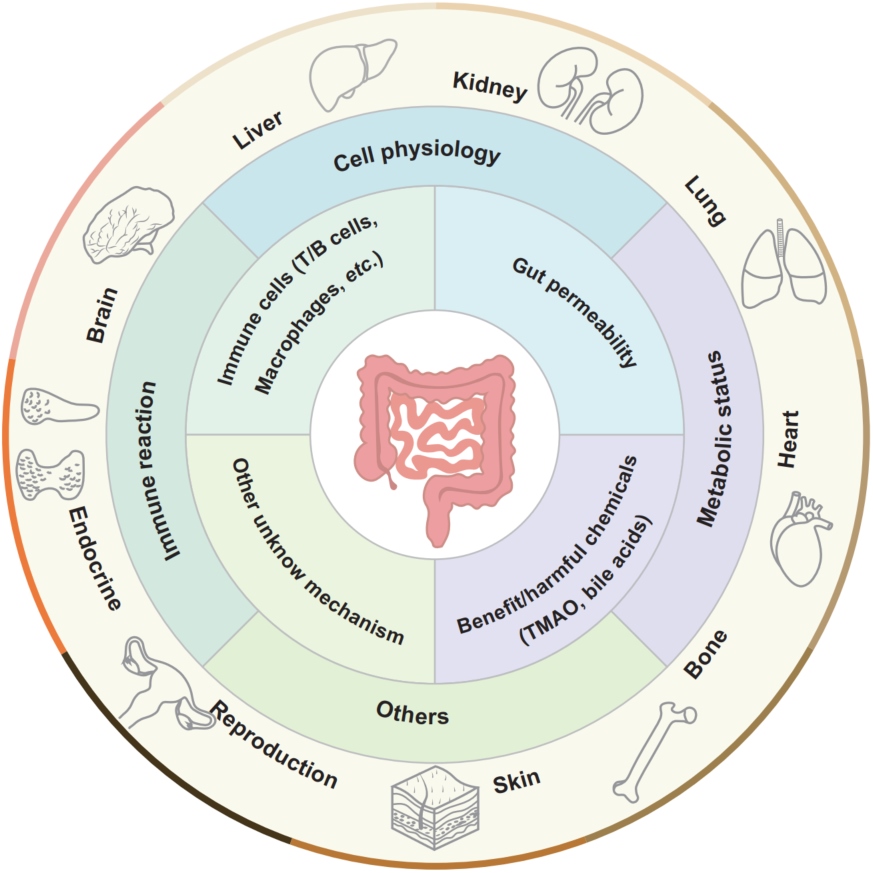
The concept of “gut–X axis”: the intestine and intestinal microbiota are proven to be able to modulate the pathophysiologic progressions of the extraintestinal organs' diseases. The bioactive chemicals and/or intestinal immune cells can translocate into the circulatory system and other organs and influence the immune reactions, metabolic status, cells physiology, and so forth of extraintestinal organs, finally regulating these organs' homeostasis. Meanwhile, other organs may reversely impact the intestine, namely such regulatory axis is bidirectional.
Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp
- 08 May 2023
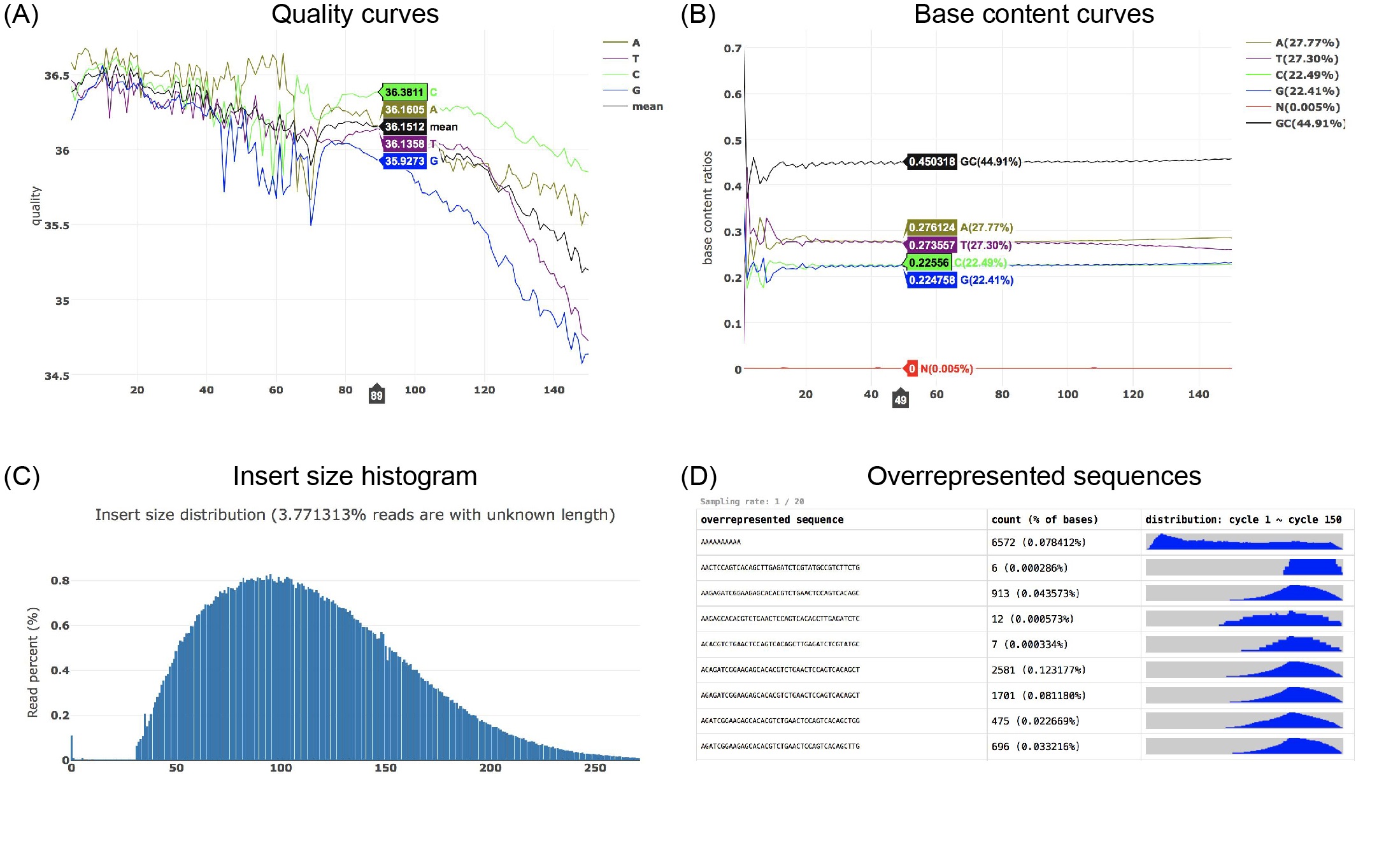
Fastp is a widely adopted tool for FASTQ data preprocessing and quality control. It is ultrafast and versatile and can perform adapter removal, global or quality trimming, read filtering, unique molecular identifier processing, base correction, and many other actions within a single pass of data scanning. Fastp has been reconstructed and upgraded with some new features. Compared to fastp 0.20.0, the new fastp 0.23.2 is even 80% faster.
ImageGP: An easy‐to‐use data visualization web server for scientific researchers
- 21 February 2022
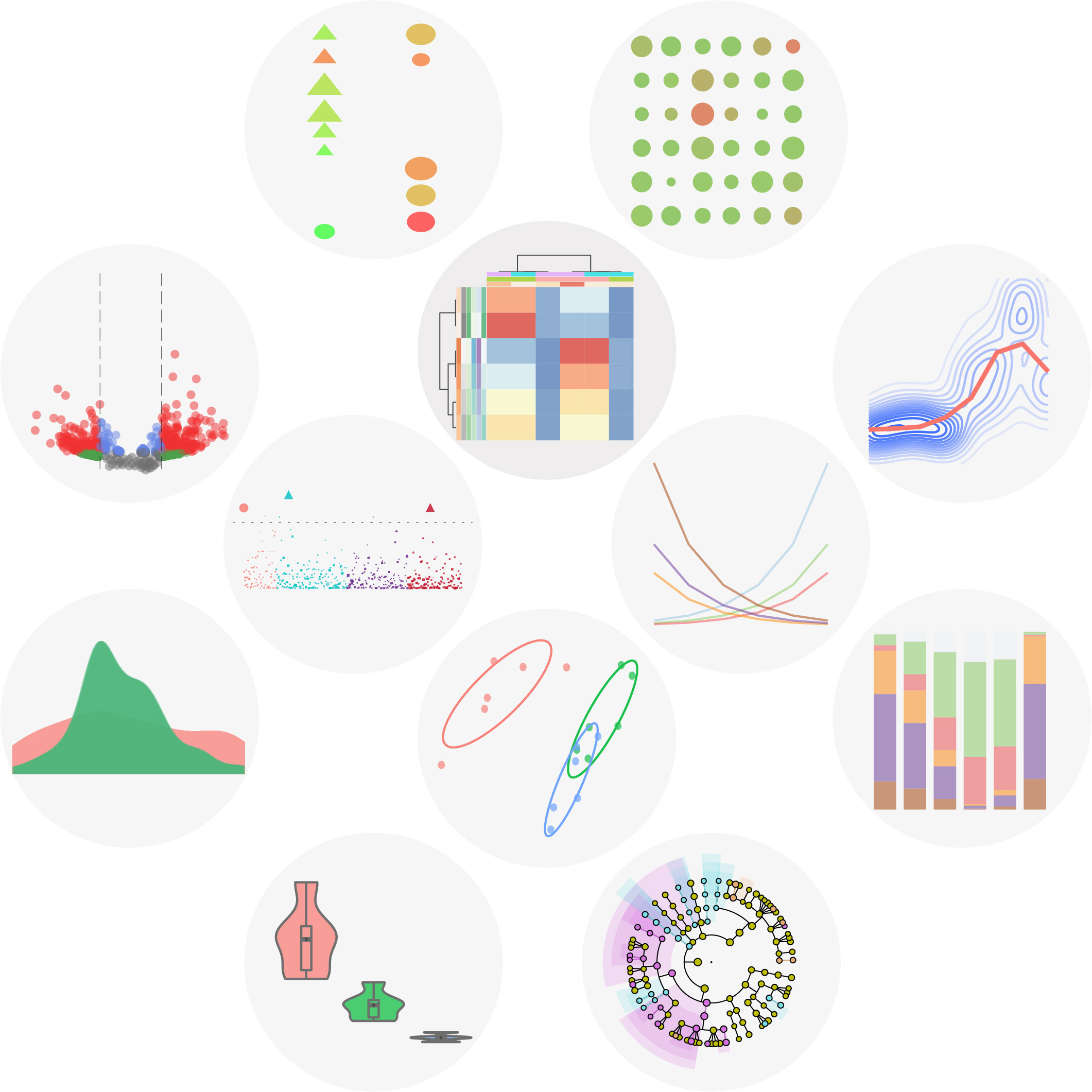
Representative visualization results of ImageGP. ImageGP supports 16 types of images and four types of online analysis with up to 26 parameters for customization. ImageGP also contains specialized plots like volcano plot, functional enrichment plot for most omics-data analysis, and other 4 specialized functions for microbiome analysis. Since 2017, ImageGP has been running for nearly 5 years and serving 336,951 visits from all over the world. Together, ImageGP (http://www.ehbio.com/ImageGP/) is an effective and efficient tool for experimental researchers to comprehensively visualize and interpret data generated from wet-lab and dry-lab.
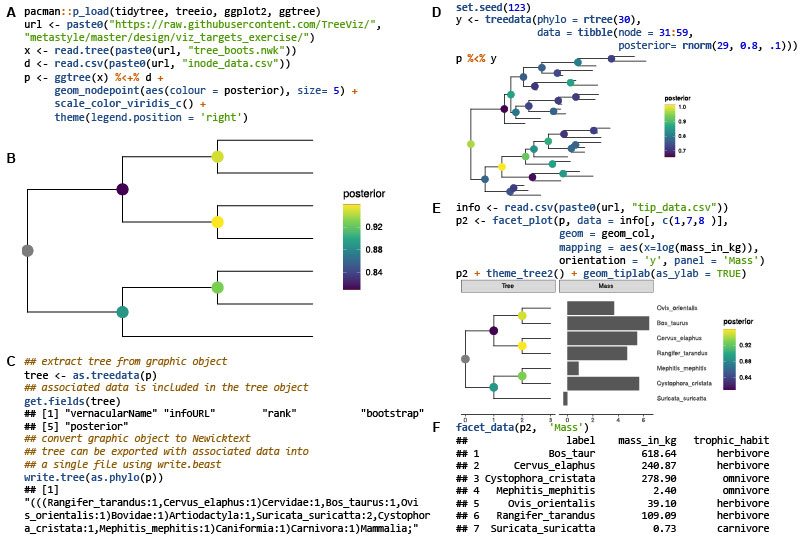
Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data
- 28 September 2022
Using PhyloSuite for molecular phylogeny and tree-based analyses
- 16 February 2023
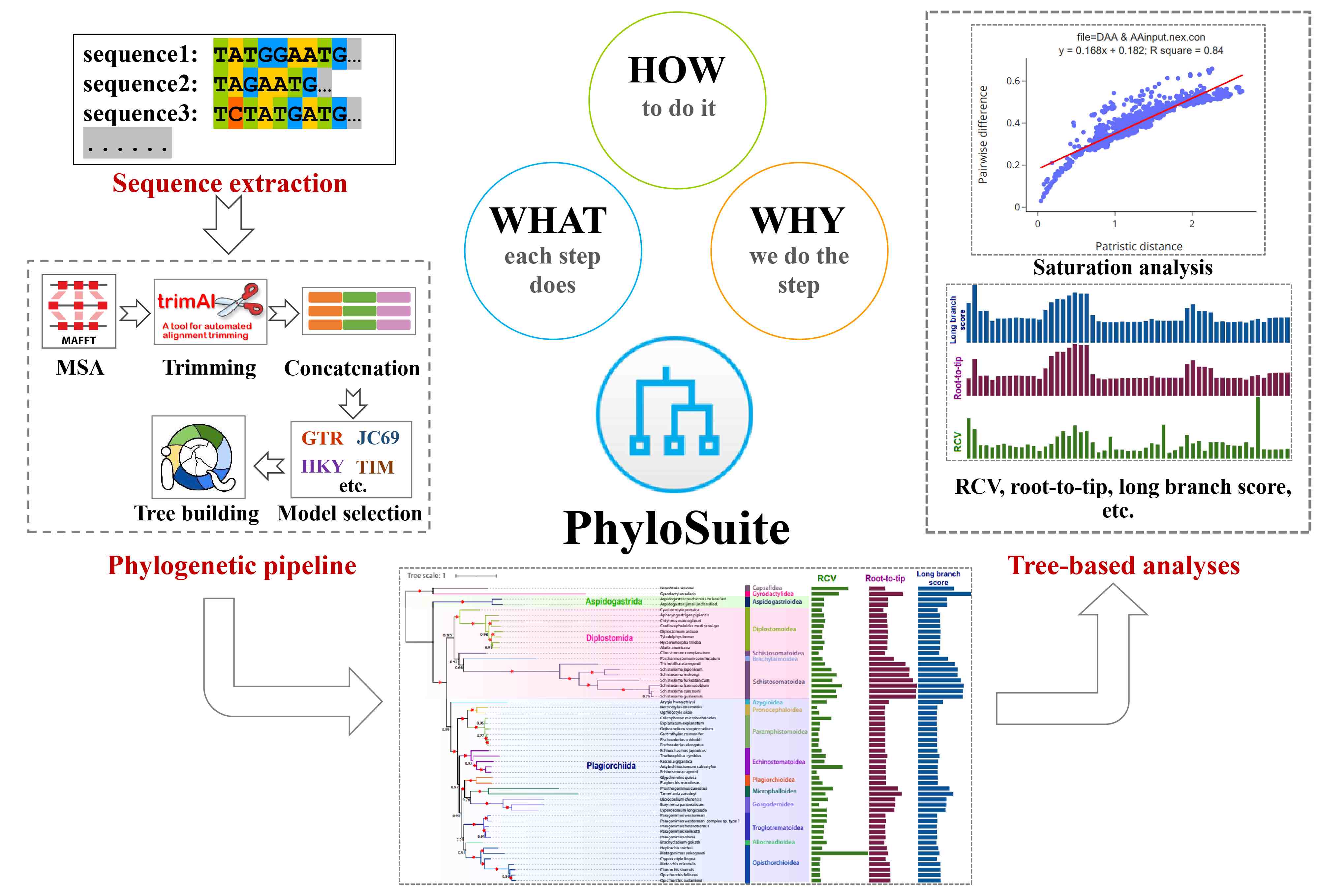
A new release of PhyloSuite, capable of conducting tree-based analyses. Detailed guidelines for each step of phylogenetic and tree-based analyses, following the “What? Why? and How?” structure. This protocol will help beginners learn how to conduct multilocus phylogenetic analyses and help experienced scientists improve their efficiency.