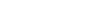Multiomics analysis reveals signatures of selection and loci associated with complex traits in pigs
- 15 December 2024
By combining nine high-quality pig genomes and 1081 resequencing data sets, researchers constructed a comprehensive pan-genome and genetic variation data set for pigs, laying the groundwork for analyzing complex traits. This data set identified selective sweep signals and genes related to fat deposition, muscle growth, and immune function, providing genetic markers for breeding. Integration of transcriptome and methylome data from 27 muscle development stages revealed epigenetic mechanisms behind muscle growth differences between Eastern and Western pig breeds. Artificial selection influenced DNA methylation and gene regulation, affecting muscle growth and meat quality. Key genes, such as GHSR and BDH1, regulate skeletal muscle development and meat production, while ABCA3 impacts immune response and lung injury, and BRCA1 is linked to adipocyte growth and fat deposition.
MetOrigin 2.0: Advancing the discovery of microbial metabolites and their origins
- 06 November 2024
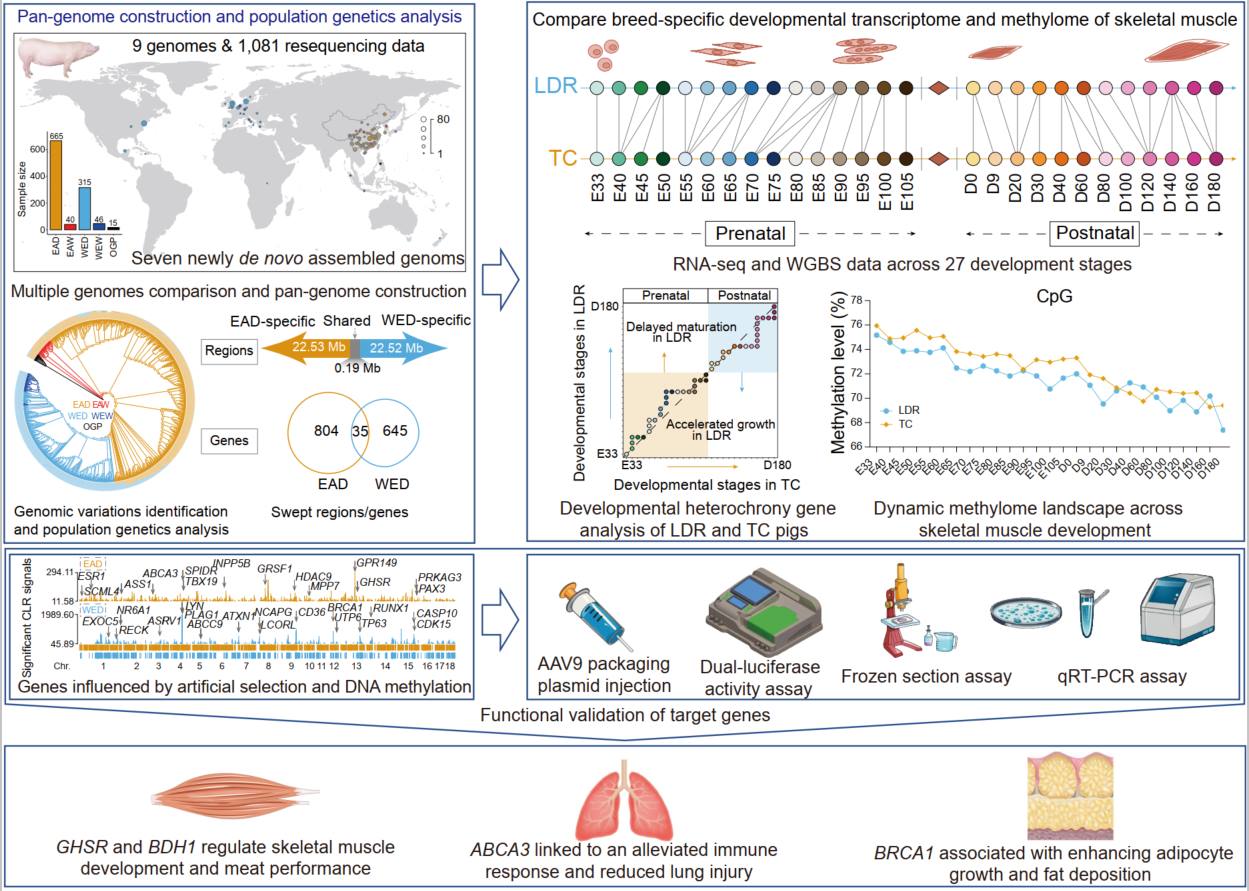
MetOrigin serves as a powerful web-based tool for microbiome-metabolome data integration, helping researchers pinpoint microbial metabolites and trace their origins. The newly upgraded MetOrigin 2.0 comes equipped with three analytical modes and eight enhanced/new modules for more precise microbial metabolite identification, including quick database search, origin analysis, pathway analysis, Sankey network, correlation analysis, orthology analysis, mediation analysis, and network visualization. The backend database has been updated with the latest data from seven key public databases (KEGG, HMDB, BIGG, ChEBI, FoodDB, Drugbank, and T3DB), with expanded coverage of 210,732 metabolites, each linked to its source organism.
Bioprospecting of culturable marine biofilm bacteria for novel antimicrobial peptides
- 17 October 2024
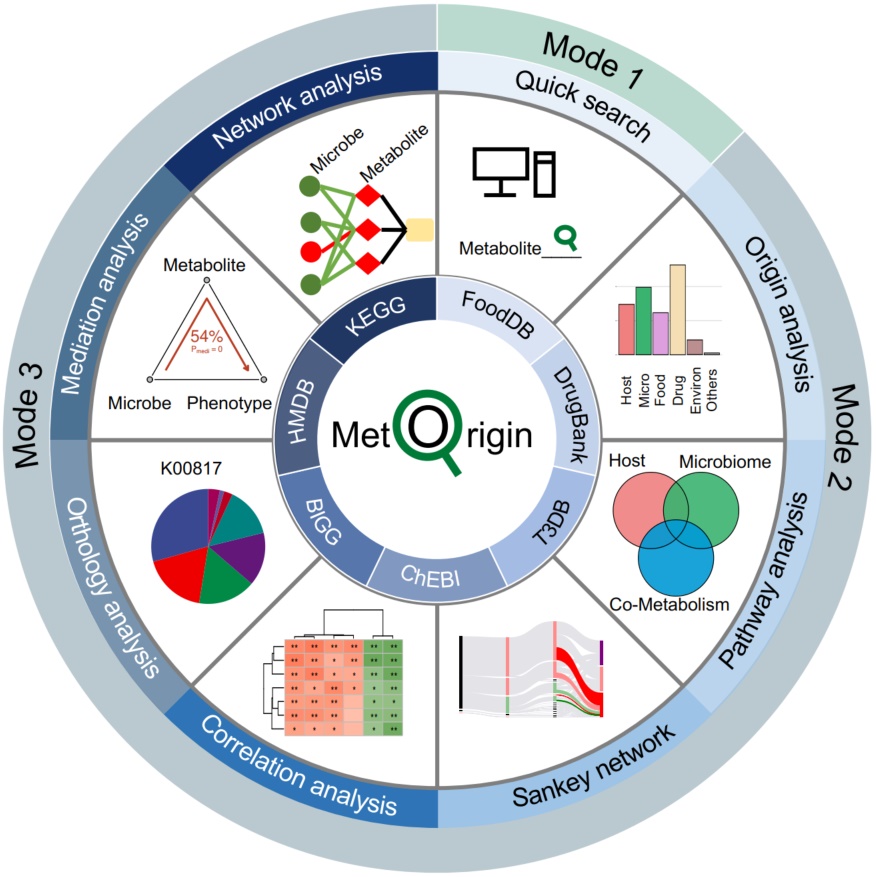
In the context of marine resource bioprospecting, we construct a bank of culturable marine biofilm bacteria encompassing 713 strains and their nearly complete genomes. Using ribosome profiling and a new deep learning algorithm, we get more than 300 candidate antimicrobial peptides (AMPs). We further conduct antimicrobial activity and cytotoxicity experiments to evaluate their potential as druggable molecules.
The crop mined phosphorus nutrition via modifying root traits and rhizosphere micro-food web to meet the increased growth demand under elevated CO2
- 25 October 2024
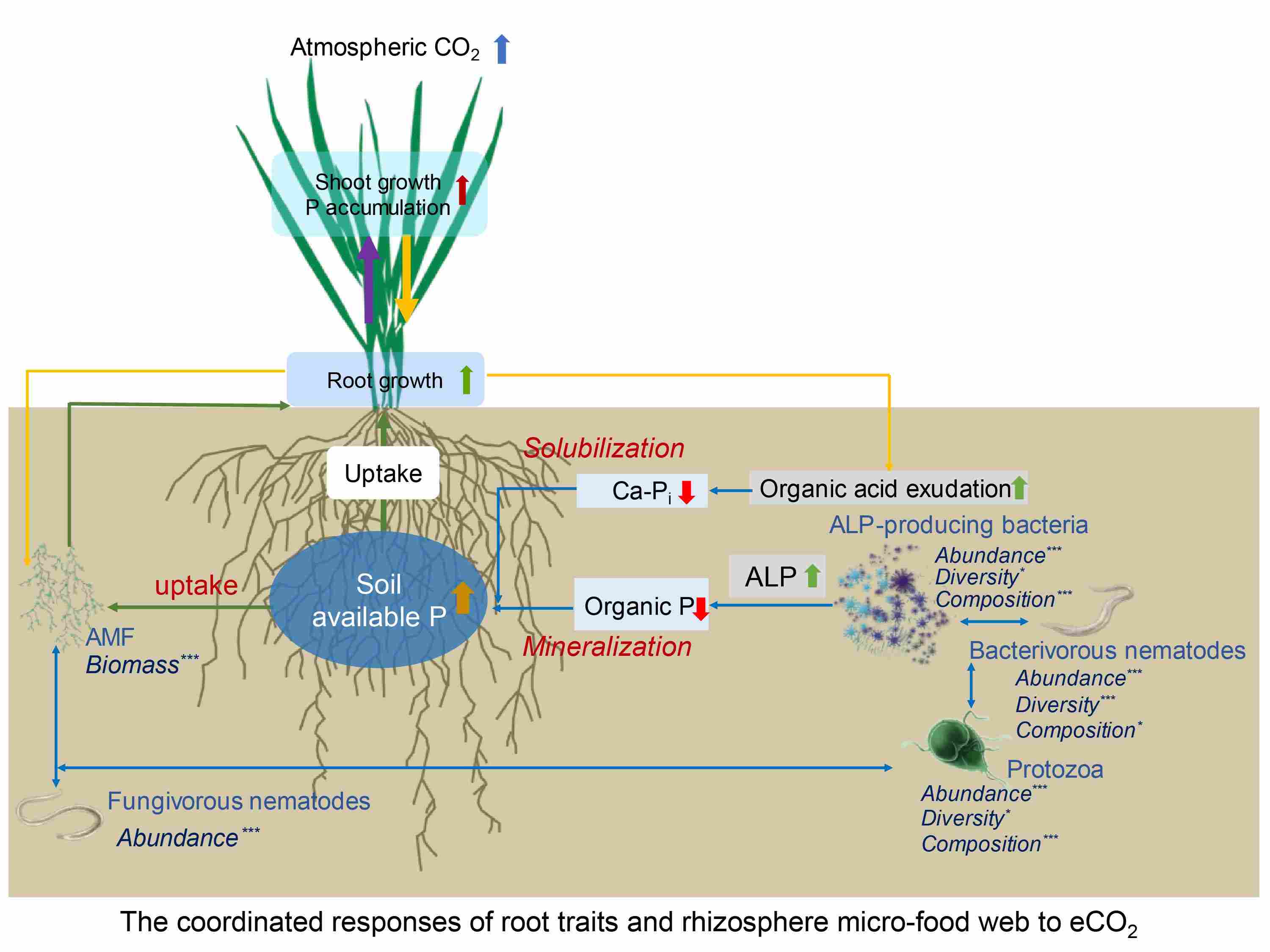
The Elevated CO2 (eCO2) promoted wheat phosphorus (P) accumulation through increased root length and arbuscular mycorrhizal fungi (AMF) hyphal biomass, the concentration of organic acid anions and the alkaline phosphomonoesterase (ALP) activity. The eCO2 also increased the growth of ALP-producing bacteria, protozoa and bacterivorous and fungivorous nematodes in the rhizosphere, and strengthened their trophic interactions.
Comprehensive multi-tissue epigenome atlas in sheep: A resource for complex traits, domestication, and breeding
- 15 December 2024
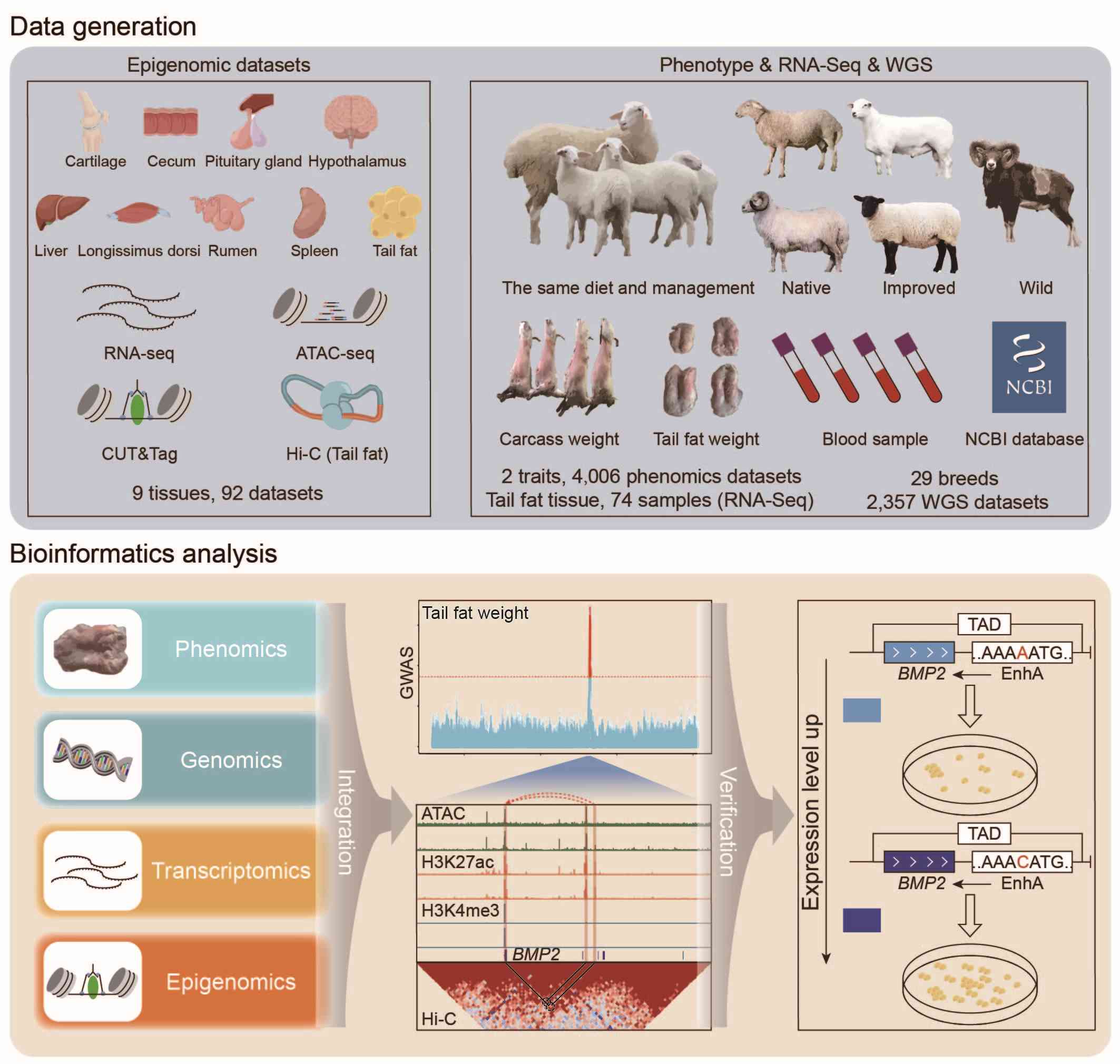
We constructed the first multi-tissue epigenome atlas in sheep, identifying a total of 753,723 nonredundant functional elements, with over 60% being novel. Our findings highlight tissue-specific functional elements, particularly those related to sensory abilities and immune response, which are highly enriched in genomic regions influenced by domestication. We identified the A allele of Chr13:51760995A>C as a favorable SNP through integrated analysis of multi-omics datasets. Our study provides foundational data resources and a successful case for understanding the adaptive evolution and complex traits of sheep.
Comprehensive lung microbial gene and genome catalogs assist the mechanism survey of Mesomycoplasma hyopneumoniae strains causing pig lung lesions
- 26 December 2024
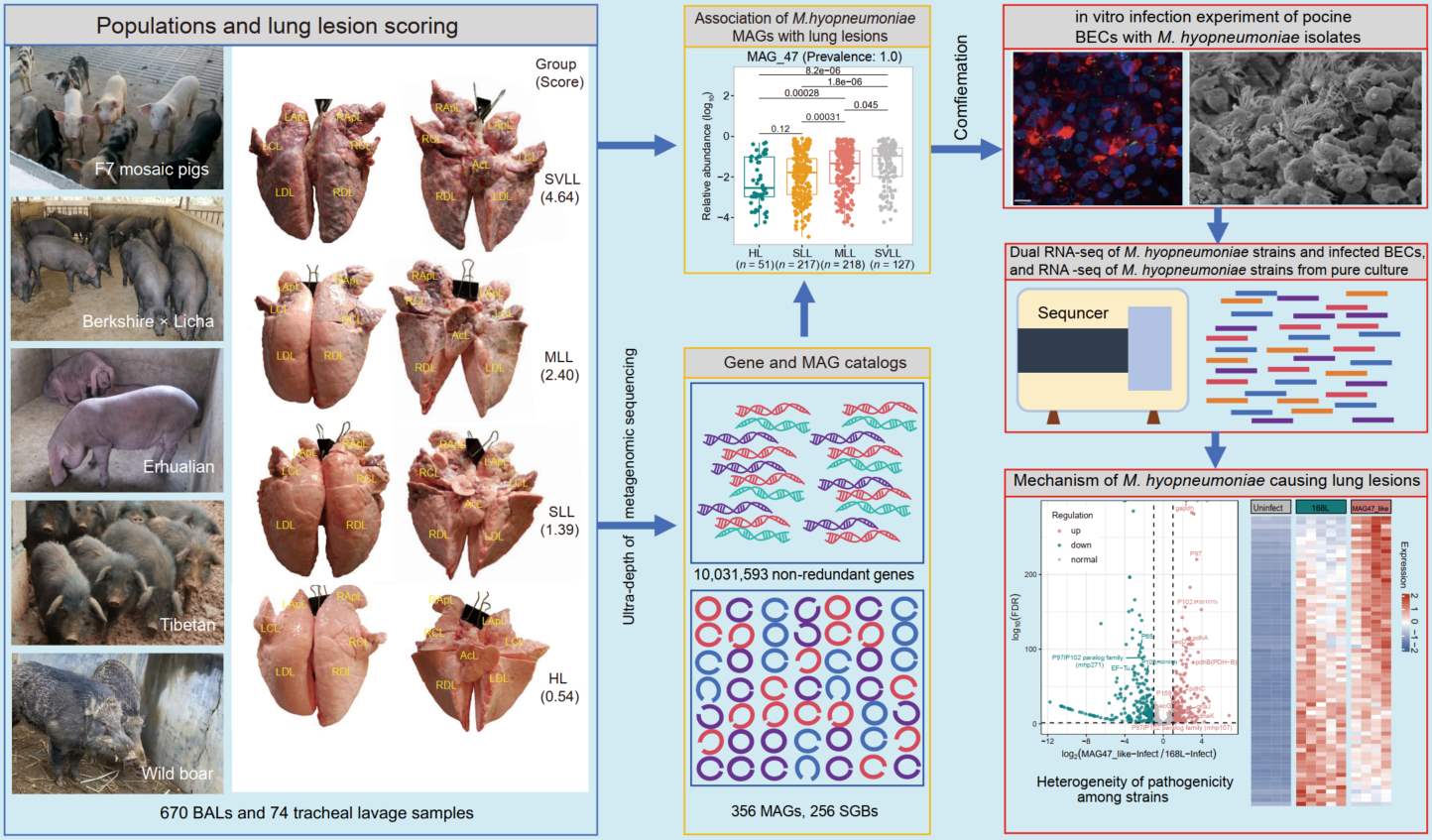
In this study, we utilized 744 metagenomic sequencing data from lower respiratory tract microbial samples of 675 well-phenotyped pigs across five cohorts. From these, we constructed a microbial gene catalog containing 10,031,593 nonredundant genes and 356 metagenome-assembled genomes (MAGs), which were further clustered into 256 species-level genome bins. Based on these data sets, we observed the significant association between Mesomycoplasma hyopneumoniae (M. hyopneumoniae) and lung lesions at the strain level. This association was confirmed by in vitro infection of porcine bronchial epithelial cells (BECs) with two M. hyopneumoniae strains. By employing dual RNA sequencing of both M. hyopneumoniae and infected BECs, alongside RNA-sequencing data from two pure-cultured M. hyopneumoniae strains, we elucidated the mechanism of M. hyopneumoniae causing pig lung lesions and explained the observed heterogeneity in pathogenicity among M. hyopneumoniae strains.
Gut Bifidobacterium pseudocatenulatum protects against fat deposition by enhancing secondary bile acid biosynthesis
- 20 December 2024
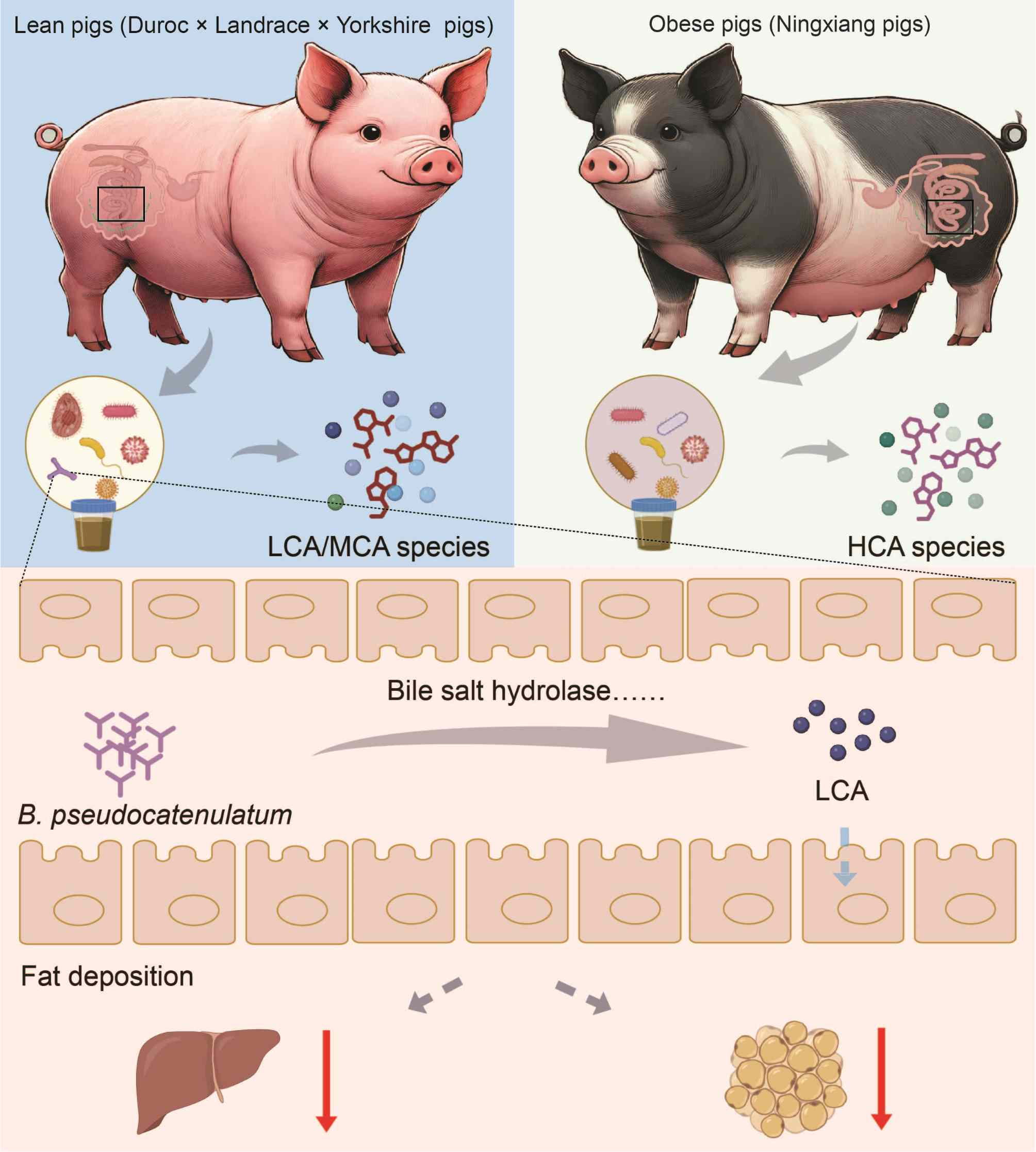
This figure highlights how gut microbiota and bile acids regulate fat deposition in lean and obese pigs. Lean pigs showed increased microbial-derived lithocholic acid (LCA) species and muricholic acid species, closely linked to the abundance of Bifidobacterium pseudocatenulatum. In contrast, obese pigs exhibited gut microbiota favoring hyodeoxycholic acid species accumulation. In high-fat diet-fed mice, live B. pseudocatenulatum supplementation enhances secondary bile acid synthesis, especially LCA production, and reduces fat mass, while bile salt hydrolase (BSH) inhibition negates these effects. Dietary LCA similarly lowers fat deposition in obese pigs and rats. These findings provide mechanistic insights into the anti-fat deposition role of B. pseudocatenulatum and identify BSH as a potential target for preventing excessive fat deposition in humans and animals.
Pangenome and genome variation analyses of pigs unveil genomic facets for their adaptation and agronomic characteristics
- 26 December 2024
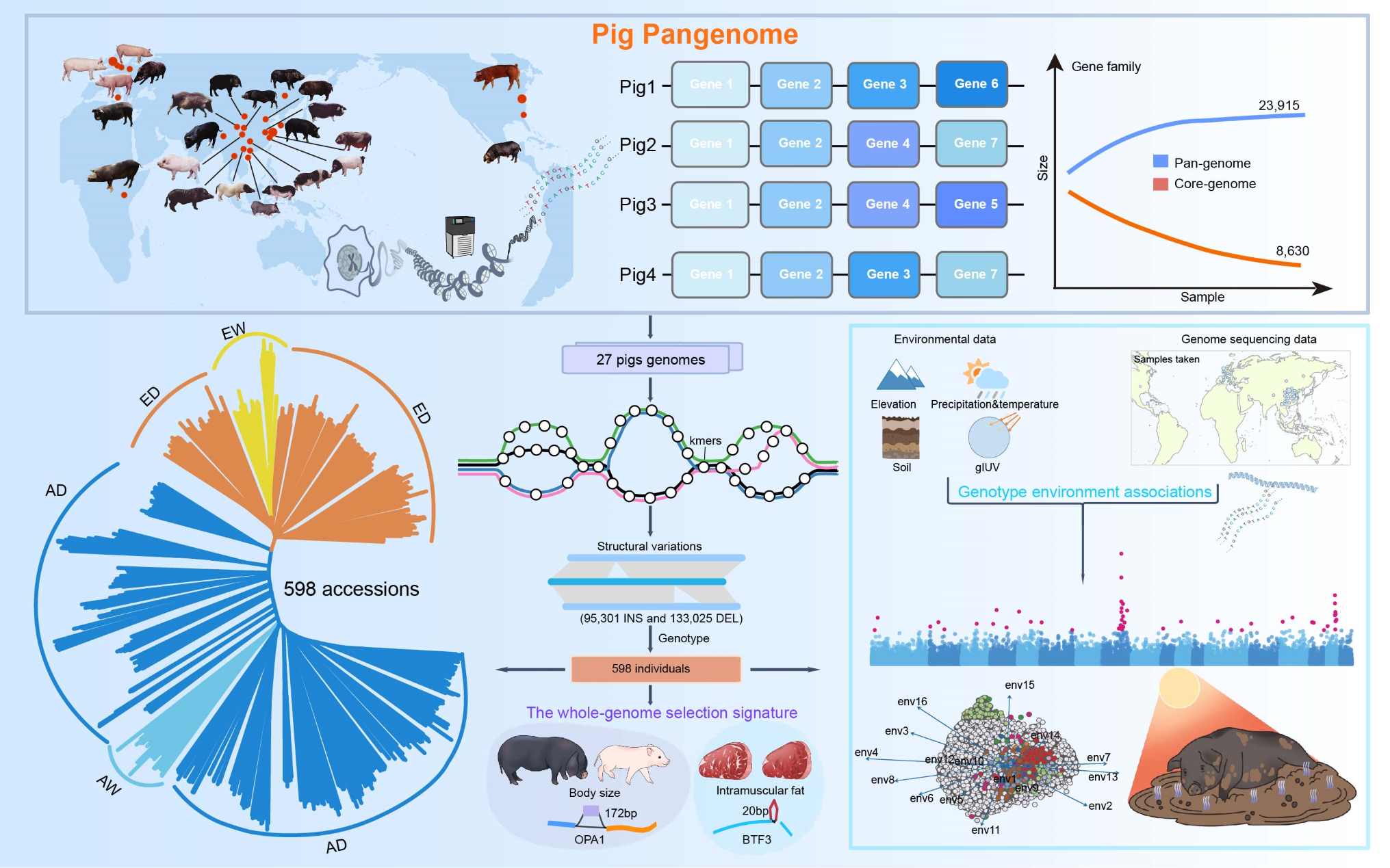
The development of a comprehensive pig graph pangenome assembly encompassing 27 genomes represents the most extensive collection of pig genomic data to date. Analysis of this pangenome reveals the critical role of structural variations in driving adaptation and defining breed-specific traits. Notably, the study identifies BTF3 as a key candidate gene governing intramuscular fat deposition and meat quality in pigs. These findings underscore the power of pangenome approaches in uncovering novel genomic features underlying economically important agricultural traits. Collectively, these results demonstrate the value of leveraging large-scale, multi-genome analyses for advancing our understanding of livestock genomes and accelerating genetic improvement.
Targeting SLC7A11/xCT improves radiofrequency ablation efficacy of HCC by dendritic cells mediated anti-tumor immune response
- 20 November 2024
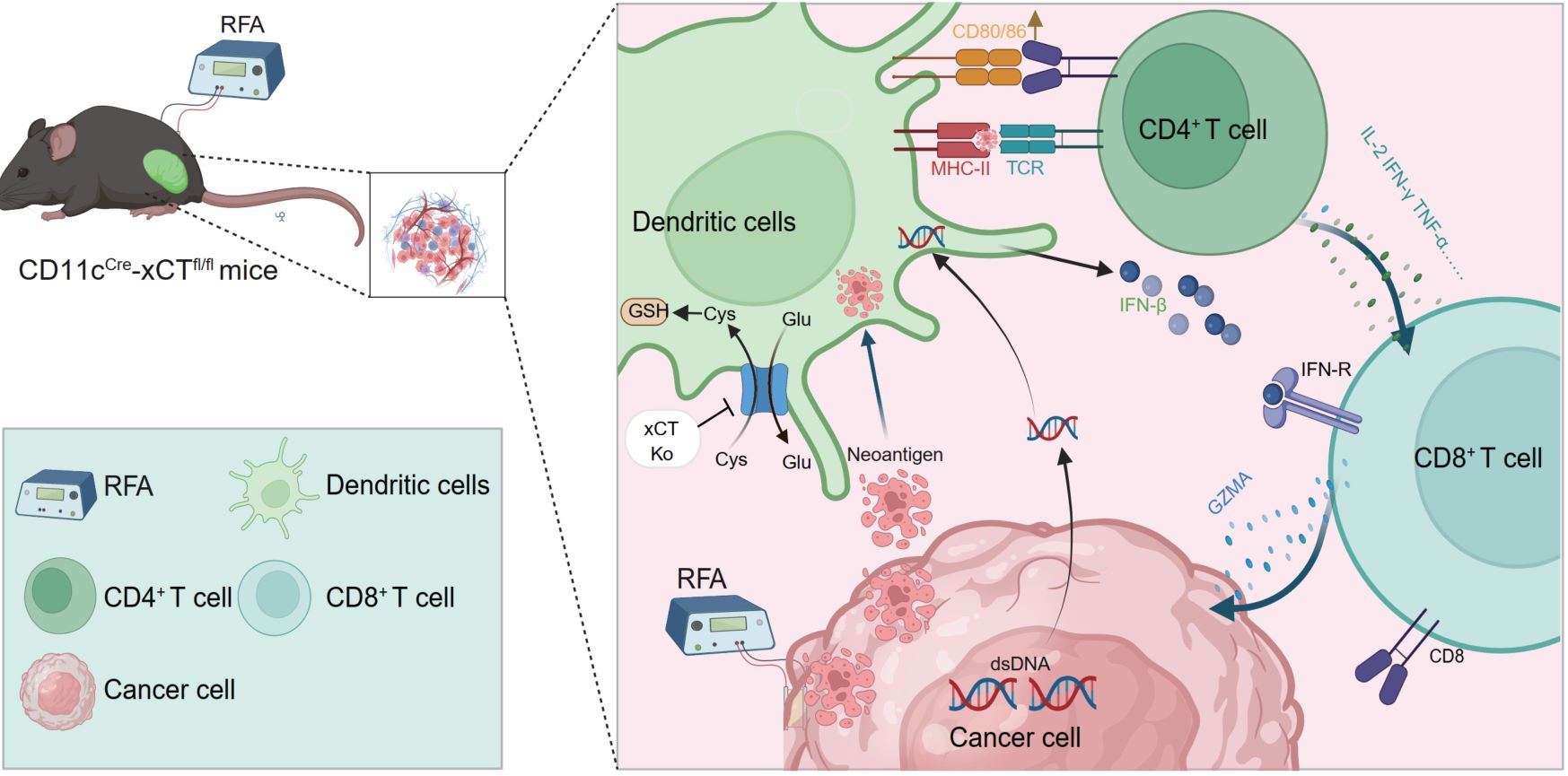
After RFA treatment in patients with liver cancer, the expression of SLC7A11/xCT and the proportion of DCs in the TME were significantly increased. SLC7A11/xCT is a poor prognostic marker for liver cancer and is mainly expressed in DCs in the TME. Targeting xCT in DCs combined with RFA significantly enhances anti-tumor immunity, suppressing tumor growth and offering a promising strategy for improved therapeutic outcomes in liver cancer.
Metagenomic analysis sheds light on the mixotrophic lifestyle of bacterial phylum Zhuqueibacterota
- 23 November 2024
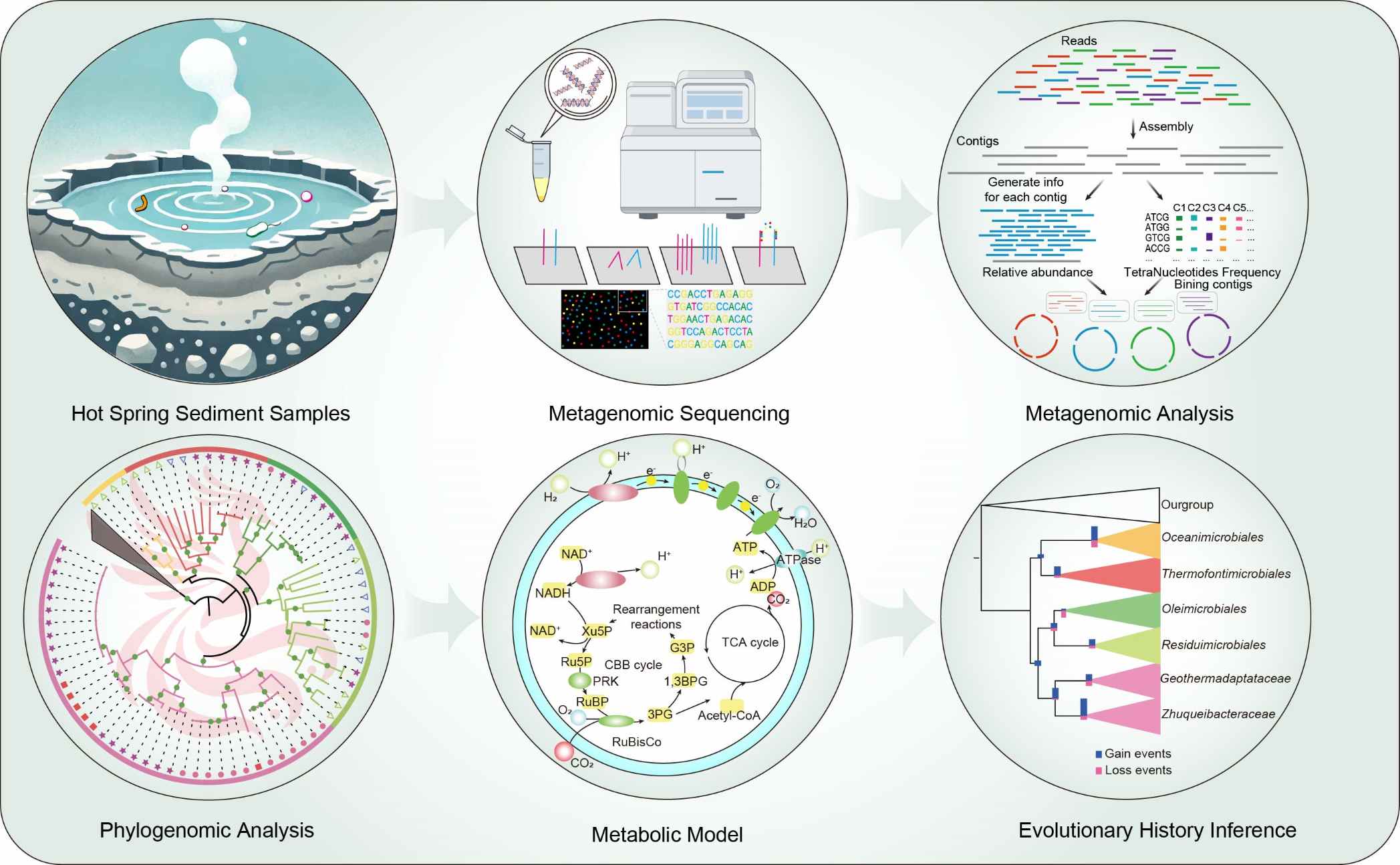
Zhuqueibacterota is a novel bacterial phylum proposed based on hot spring metagenomes and public metagenome-assembled genomes, classified within the Fibrobacterota-Chlorobiota-Bacteroidota superphylum. This globally distributed phylum consists of one class and five orders, with the majority of its members being facultative anaerobes. Notably, the order Zhuqueibacterales utilizes hydrogen as an electron donor for carbon fixation through the Calvin Benson Bassham cycle. Phylogenetic and metabolic analyses reveal the phylum's key role in the carbon cycle, with frequent horizontal gene transfer events influencing its evolutionary trajectory.
Polyploidy drives autophagy to participate in plant-specific functions
- 09 December 2024
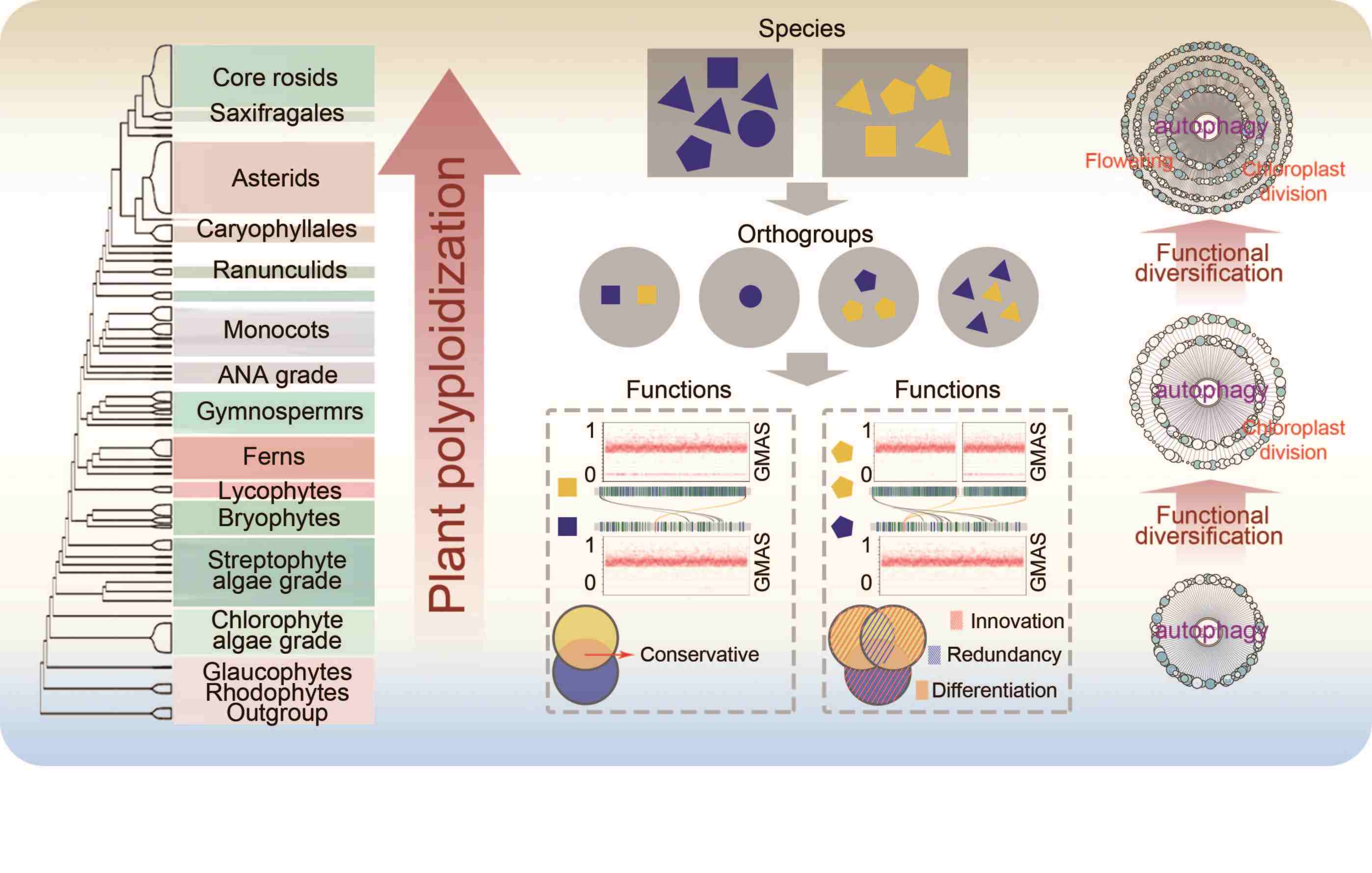
Polyploidization promotes the functional diversification of autophagy in plants, expanding autophagy-associated genes (AAGs) to support processes like chloroplast division and flowering. Analysis of 92,967 AAGs in Arabidopsis thaliana, Solanum lycopersicum, Camellia oleifera, and 74 other plant species shows that 45.69% of AAGs are polyploidy-related, highlighting polyploidy's role in linking autophagy to plant-specific functions.
Transcriptome-wide association identifies KLC1 as a regulator of mitophagy in non-syndromic cleft lip with or without palate
- 20 December 2024
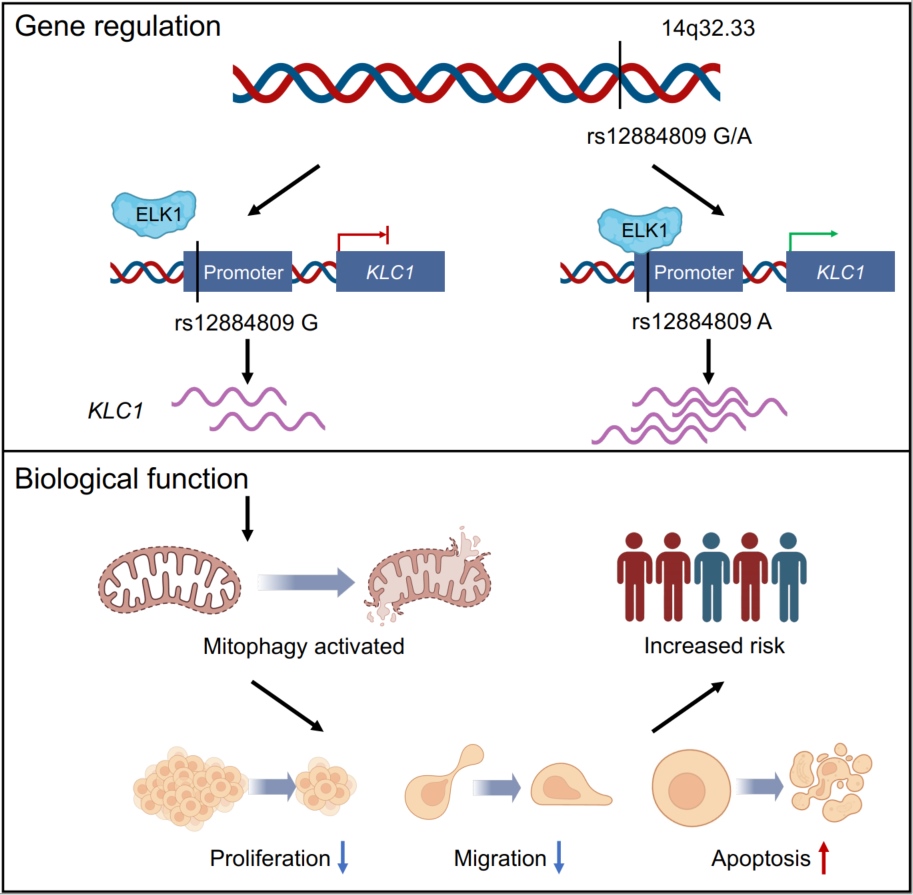
This study investigated pathogenic genes associated with non-syndromic cleft lip with or without cleft palate (NSCL/P) through transcriptome-wide association studies (TWAS). By integrating expression quantitative trait loci (eQTL) data with genome-wide association study (GWAS) data, we identified key susceptibility genes, including KLC1. Notably, the variant rs12884809 G>A was associated with an increased risk of NSCL/P by enhancing the binding of the transcription factor ELK1 to the KLC1 promoter, thereby activating its expression. This alteration in KLC1 expression subsequently impacted mitophagy, leading to significant changes in cellular behavior and zebrafish morphology. Our findings illuminate the genetic mechanisms underlying NSCL/P and provide valuable insights for future prevention strategies and a deeper understanding of this condition.
A comprehensive atlas of endogenous peptides in maize
- 11 November 2024
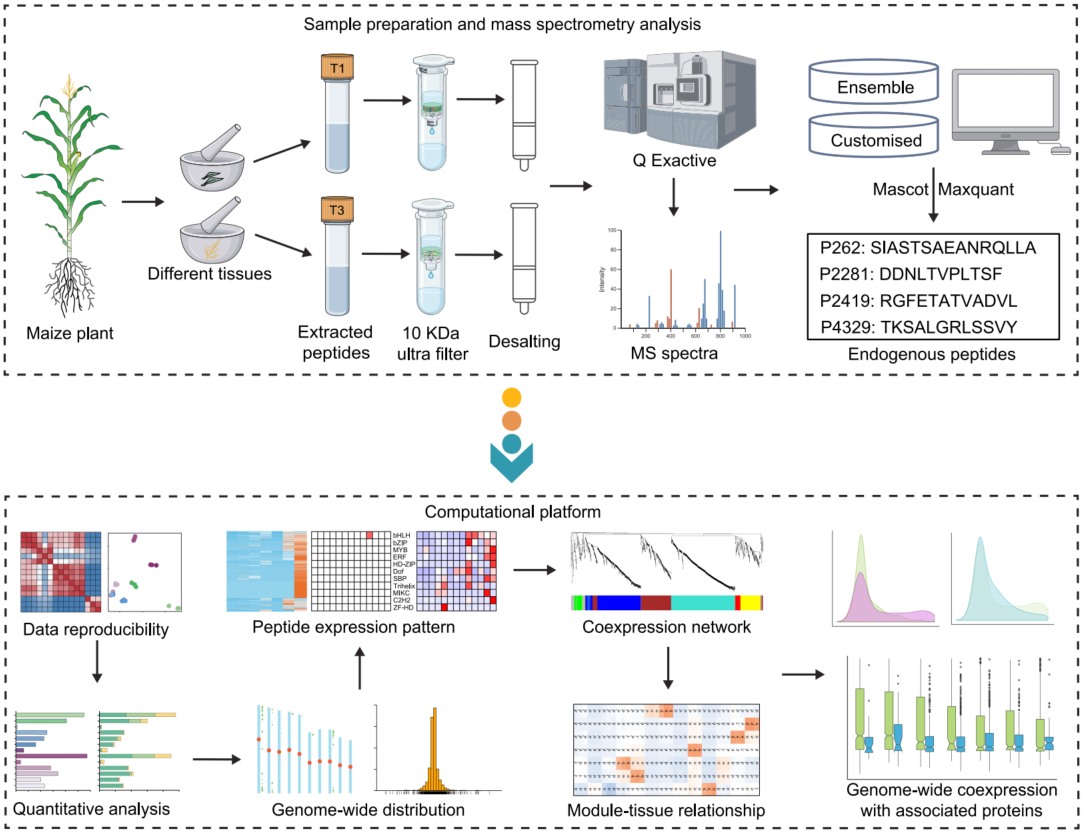
In this study, we present a comprehensive peptidomic atlas of 13 maize tissues, covering both vegetative and reproductive phases. Using a three-frame translation of canonical coding sequences, we identified 6100 nonredundant endogenous peptides, significantly expanding the known plant peptide repertoire. By integrating peptidomic coexpression profiles with previously reported proteomic profiles, we found that the peptide abundance did not consistently correlate with the abundance of their source proteins, suggesting the presence of complex regulatory mechanisms. This integrated peptidomic and proteomic map can serve as a valuable resource for exploring the functional roles of endogenous peptides in maize development and facilitates the investigation of the functional relationship among genes, peptides, and proteins across various biological contexts.
Short-term probiotic supplementation affects the diversity, genetics, growth, and interactions of the native gut microbiome
- 16 December 2024
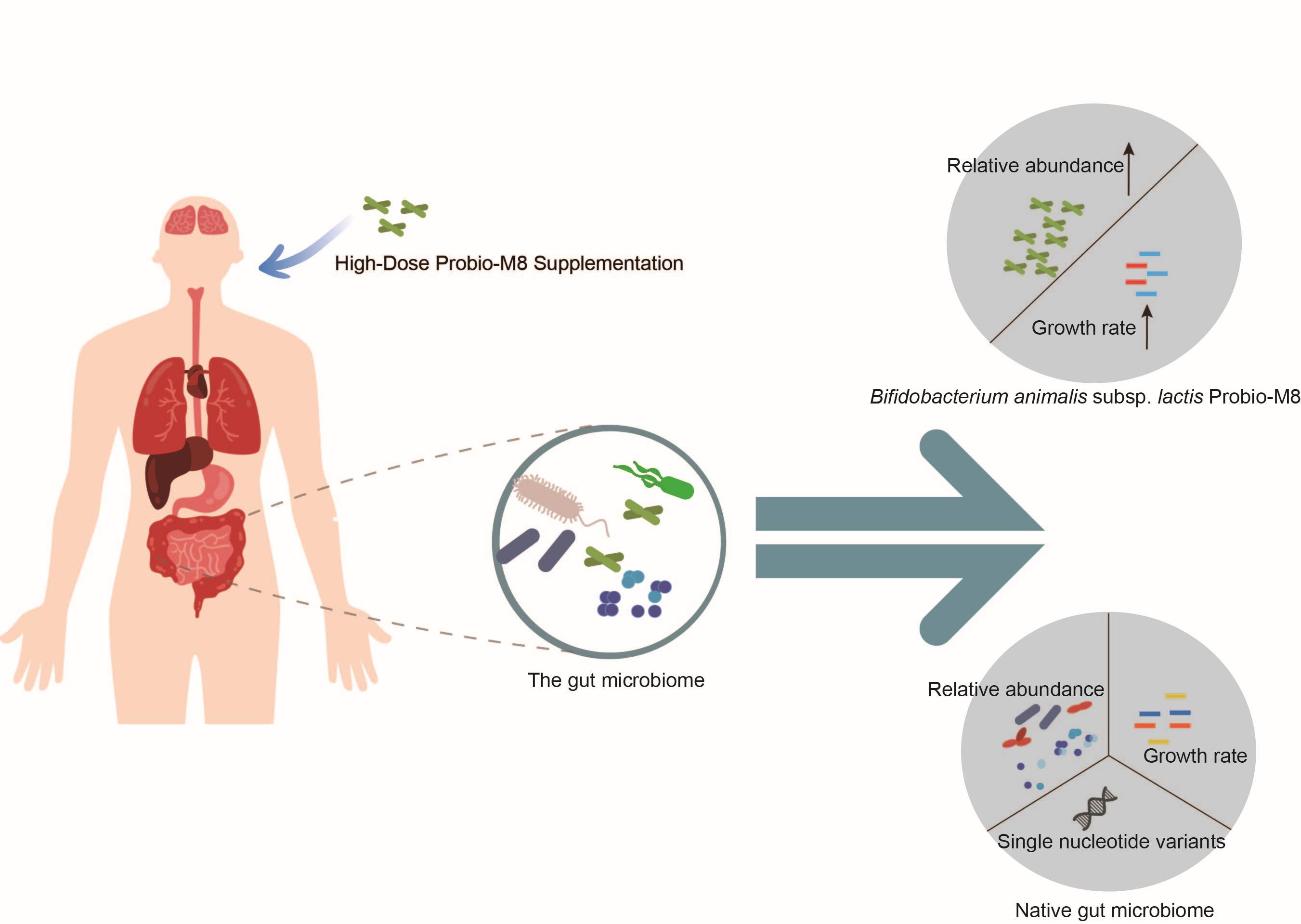
The precise mechanisms through which probiotics interact with and reshape the native gut microbiota, especially at the species and genetic levels, remain underexplored. This study employed a high-dose probiotic regimen of Bifidobacterium animalis subsp. lactis [200 billion colony forming units (CFU)/day] over 7 days among healthy participants. Weekly fecal samples were collected for metagenomic sequencing analysis. We found that probiotic intake can significantly enhance the diversity of the gut microbiome and impact single nucleotide variations, growth rates, and network interactions of the resident intestinal bacteria. These adaptive changes in the gut microbiota indicate the swift evolutionary responses of native bacteria to the ecological disturbance presented by probiotic supplementation. Notably, the microbial community appears to undergo rapid and multifaceted ecological adjustments, potentially preceding longer-term evolutionary changes. This knowledge lays the groundwork for further exploration into the mechanisms underlying probiotic-mediated modulation of the gut microbiome, highlighting the necessity of encompassing ecological and evolutionary perspectives in the design and optimization of probiotic applications.
The long-term intake of milk fat does not significantly increase the blood lipid burden in normal and high-fat diet-fed mice
- 15 December 2024
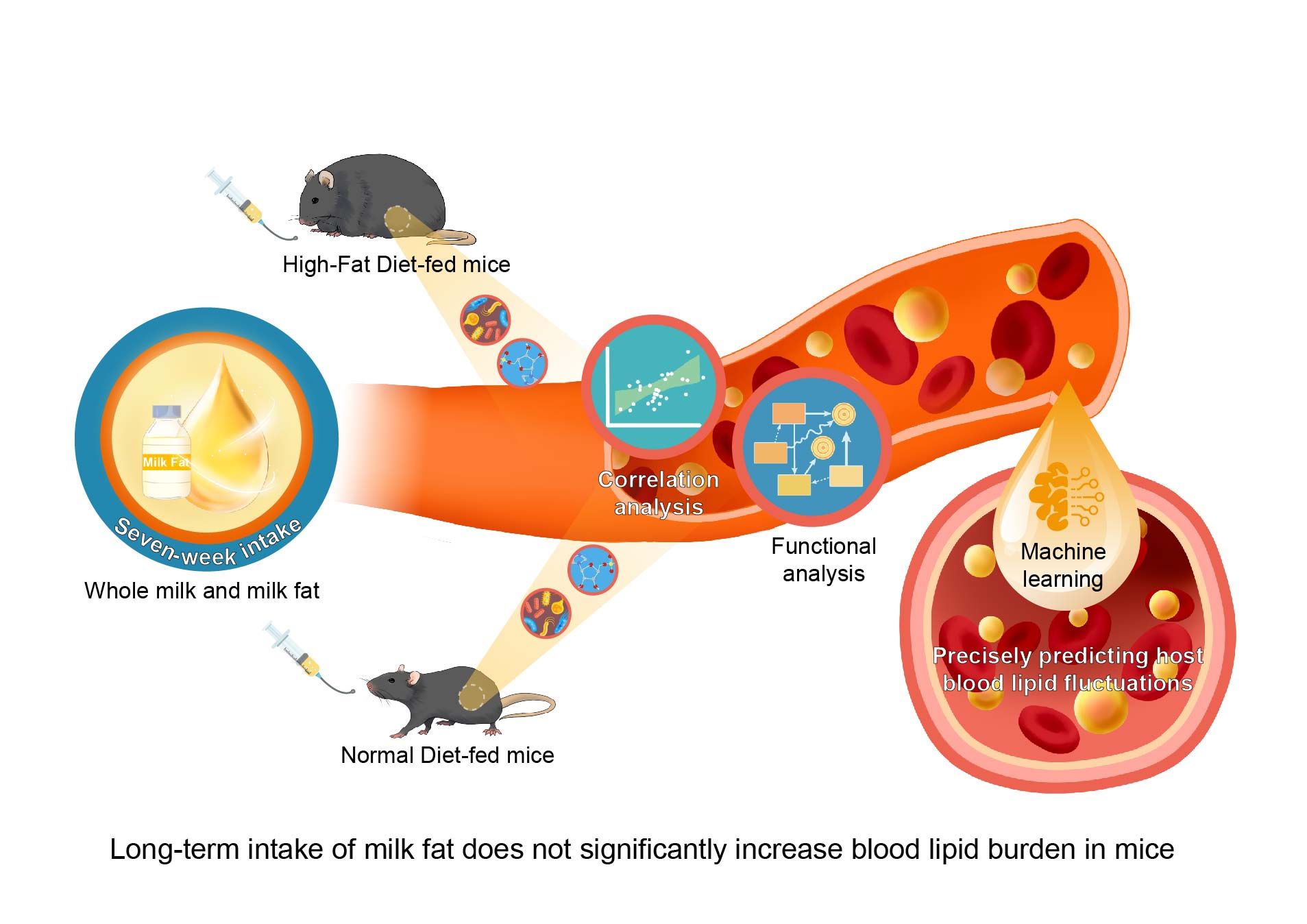
After 10 weeks of feeding C57BL/6J mice with a normal diet (ND) or a high-fat diet (HFD), a 7-week intervention with milk fat and whole milk was conducted to assess their long-term effects on host blood lipid levels. The results showed that milk fat and whole milk did not significantly elevate low-density lipoprotein cholesterol (LDL-C) in either ND- or HFD-fed mice. In ND mice, milk fat and whole milk improved gut microbiota diversity and Amplicon Sequence Variants. Key bacterial genera, such as Blautia, Romboutsia, and Prevotellaceae_NK3B31_group, were identified as bidirectional regulators of LDL-C and high-density lipoprotein cholesterol (HDL-C). Six unique metabolites were also linked to LDL-C and HDL-C regulation. Furthermore, an optimized machine learning model accurately predicted LDL-C (R² = 0.96) and HDL-C (R² = 0.89) based on gut microbiota data, with 80% of the top predictive features being gut metabolites influenced by milk fat and whole milk. These findings indicate that the long-term intake of milk fat does not significantly increase the blood lipid burden, and machine learning algorithms based on gut microbiota and metabolites offer novel insights for early lipid assessment and personalized nutrition strategies.
Unraveling the diversity dynamics and network stability of alkaline phosphomonoesterase-producing bacteria in modulating maize yield
- 20 December 2024
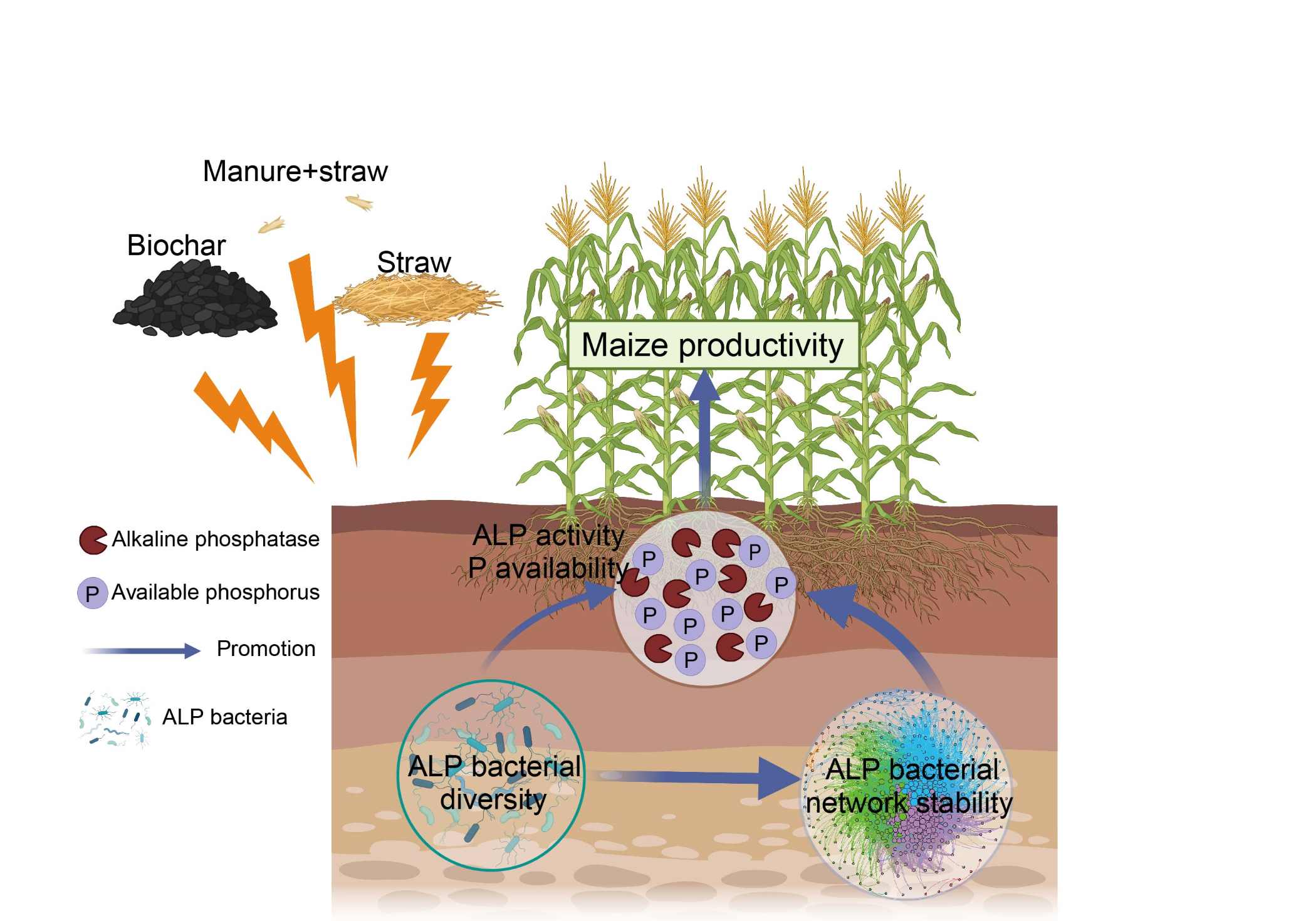
Phosphorus, as a nonrenewable resource, plays a crucial role in crop development and productivity. However, the extent to which straw amendments contribute to the dynamics of soil alkaline phosphomonoesterase (ALP)-producing bacterial community and functionality over an extended period remains elusive. Here, we conducted a 7-year long-term field experiment consisting of a no-fertilizer control, a chemical fertilizer treatment, and three straw (straw, straw combined with manure, and straw biochar) treatments. Our results indicated that straw amendments significantly improved the succession patterns of the ALP-producing bacterial diversity. Simultaneously, straw amendments significantly increased the network stability of the ALP-producing bacteria over time, as evidenced by higher network robustness, a higher ratio of negative to positive cohesion, and lower network vulnerability. High dynamic and stability of ALP-producing bacterial community generated high ALP activity which further increased soil Phosphorus (P) availability as well as maize productivity.