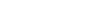ImageGP 2 for enhanced data visualization and reproducible analysis in biomedical research
- 12 September 2024
The infinity symbol illustrates the seamless workflow of ImageGP 2, encompassing essential functions such as data format transformation, data validation, and parameter combination. This process culminates in the generation of diverse visual outputs, including line, point, and bar plots. Key features include a personalized user center for managing large data sets, interactive visualizations, and streamlined error feedback mechanisms. Additionally, the introduction of the ImageGP R package enables local and batch analyses. Overall, the infinity symbol embodies the limitless potential for data analysis and visualization offered by ImageGP 2.
iNAP 2.0: Harnessing metabolic complementarity in microbial network analysis
- 23 September 2024
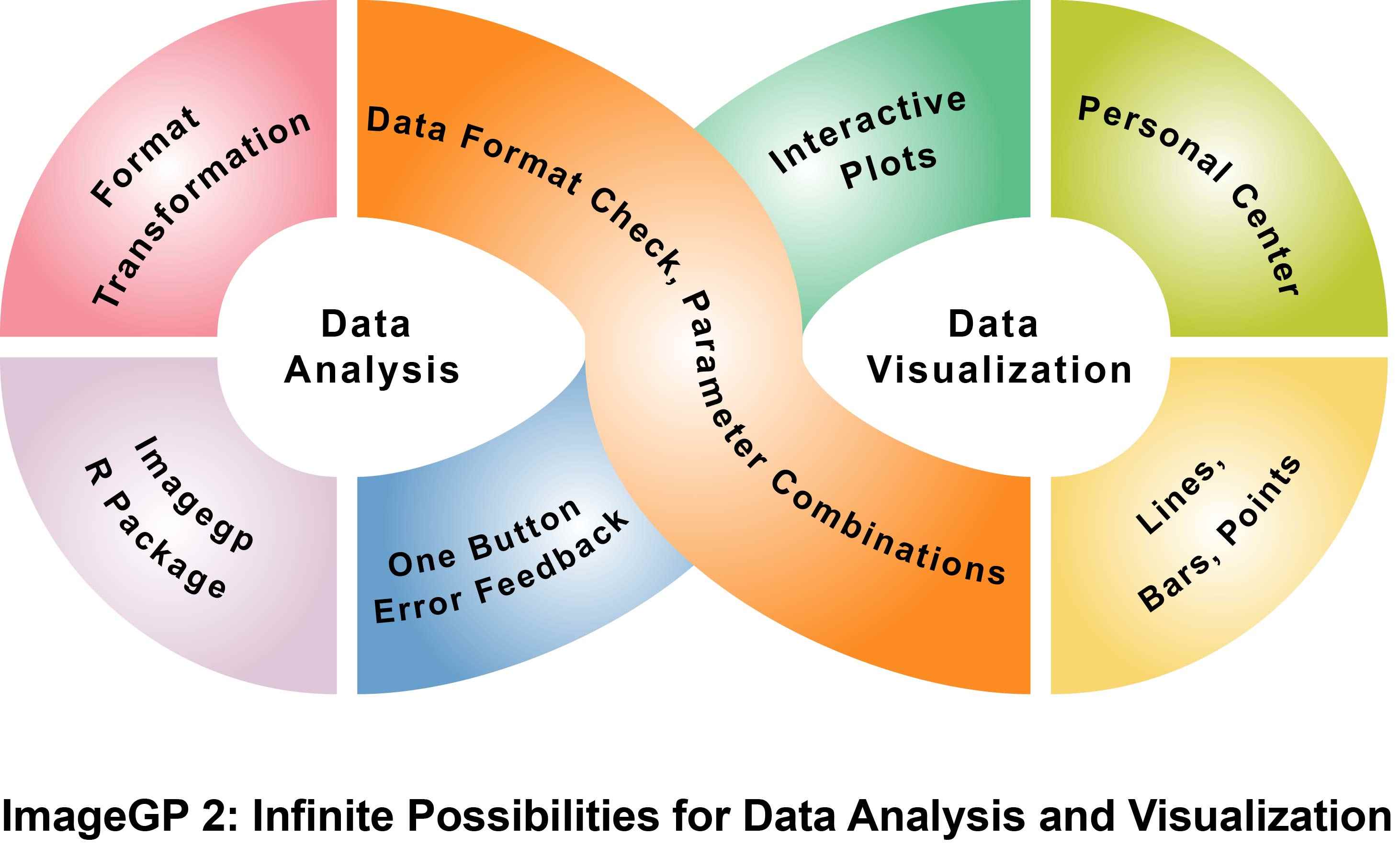
The graphical abstract illustrates the advanced capabilities of iNAP 2.0, an integrated network analysis platform designed for comprehensive metabolic interaction studies. iNAP 2.0 enables the calculation of various metabolic indices, including the PhyloMint complementarity and competition index, SMETANA (species metabolic interaction analysis) scores, and metabolic distance, facilitating in-depth analysis of metabolic complementarity and competition. It innovatively employs the random matrix theory (RMT) method to determine thresholds for constructing robust metabolic interaction networks. Additionally, the platform introduces PhyloMint PTM, which identifies potentially transferable metabolites between microbial interactions, integrating them into microbe-metabolite bipartite networks alongside traditional microbial interaction networks. This combined approach provides a holistic view of microbial metabolic exchanges, making iNAP 2.0 a powerful tool for studying microbial ecology and inferring its metabolic keystones.
Interaction between intestinal mycobiota and microbiota shapes lung inflammation
- 14 September 2024
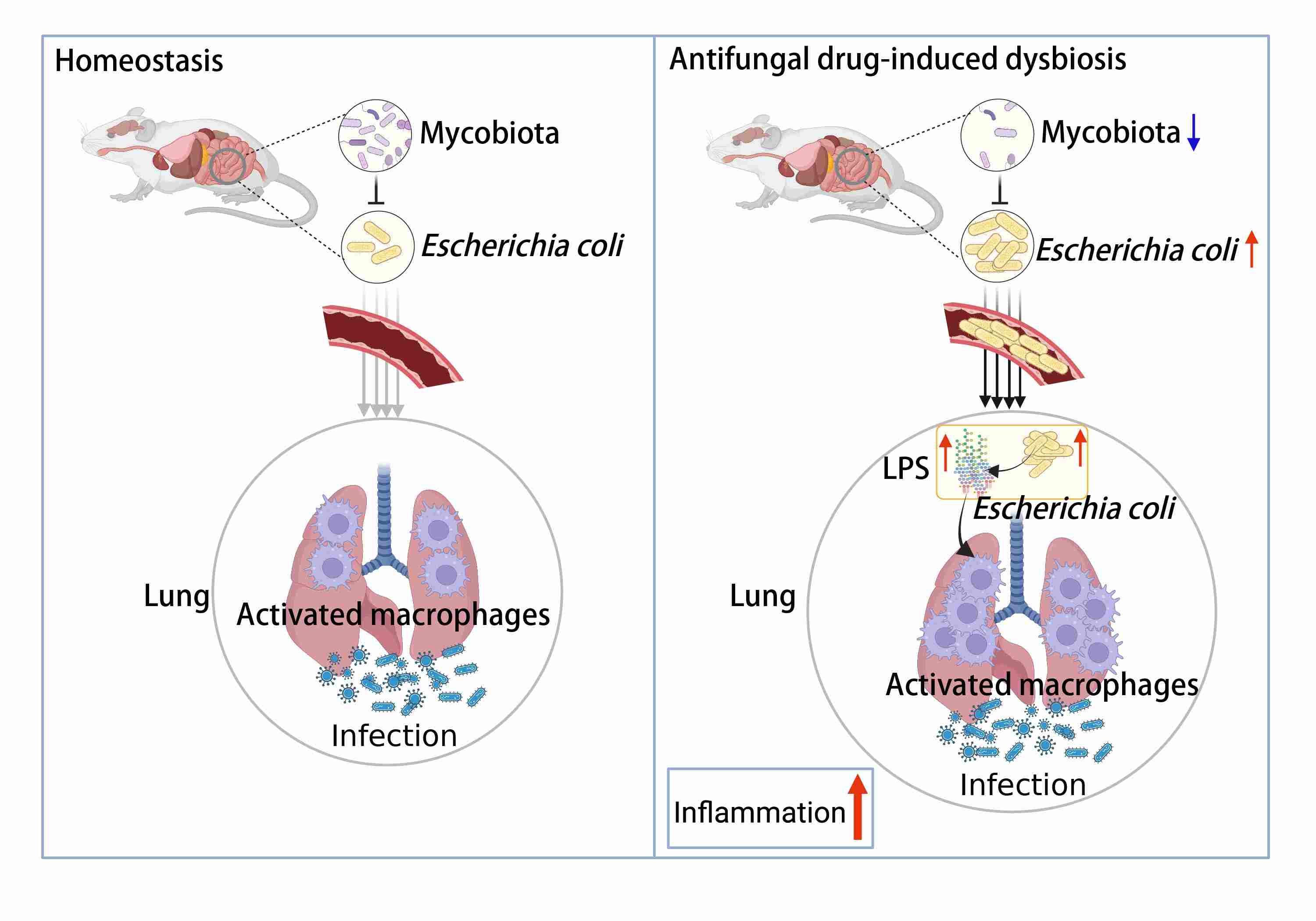
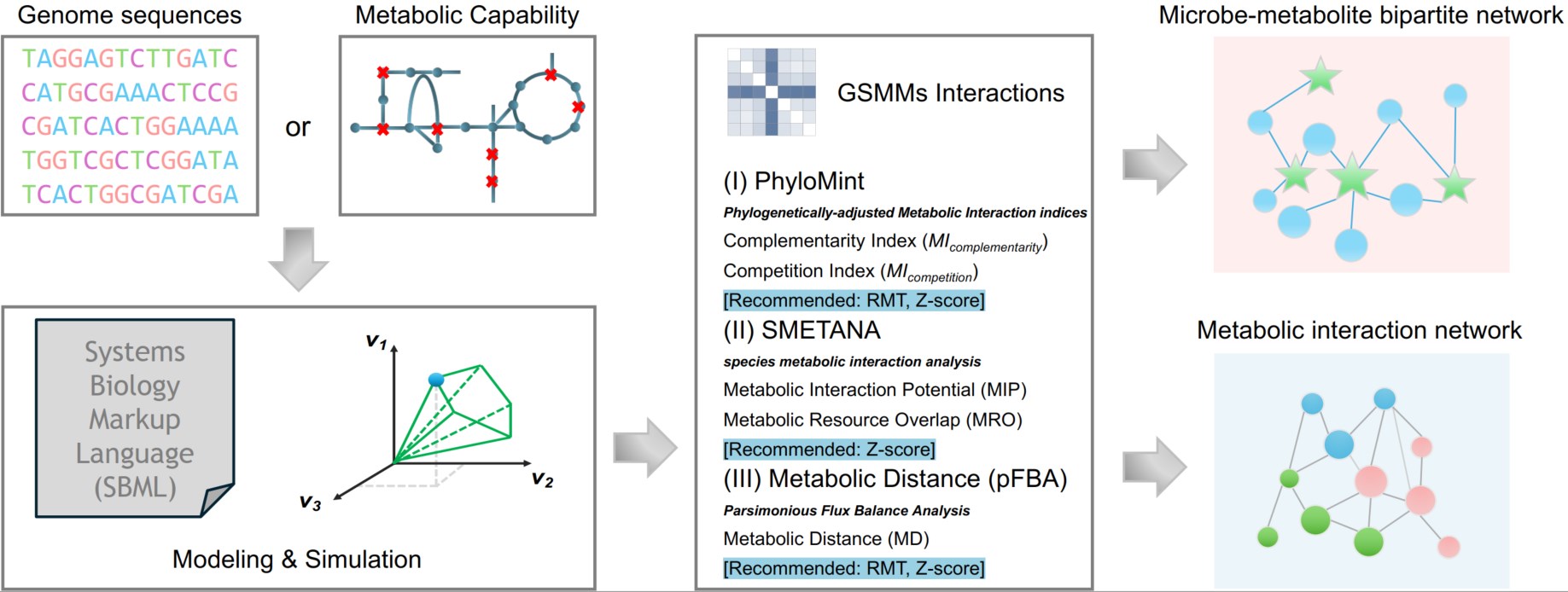
Antifungal drug-induced dysbiosis of gut mycobiota results in aggravation of lung inflammation during infection. Specifically, dysbiosis of gut mycobiota results in gut Escherichia coli overgrowth and translocation to the lung during infection. E. coli induces lung accumulation of the CD45+F4/80+Ly6G−Ly6C−CD11b+CD11c+ macrophages and activates TLR4 signaling of the CD45+F4/80+Ly6G−Ly6C−CD11b+CD11c+ macrophages. This study provided evidence for the interaction among gut inhabitants shaping the host immunity and diseases.
The rheumatoid arthritis gut microbial biobank reveals core microbial species that associate and effect on host inflammation and autoimmune responses
- 03 October 2024
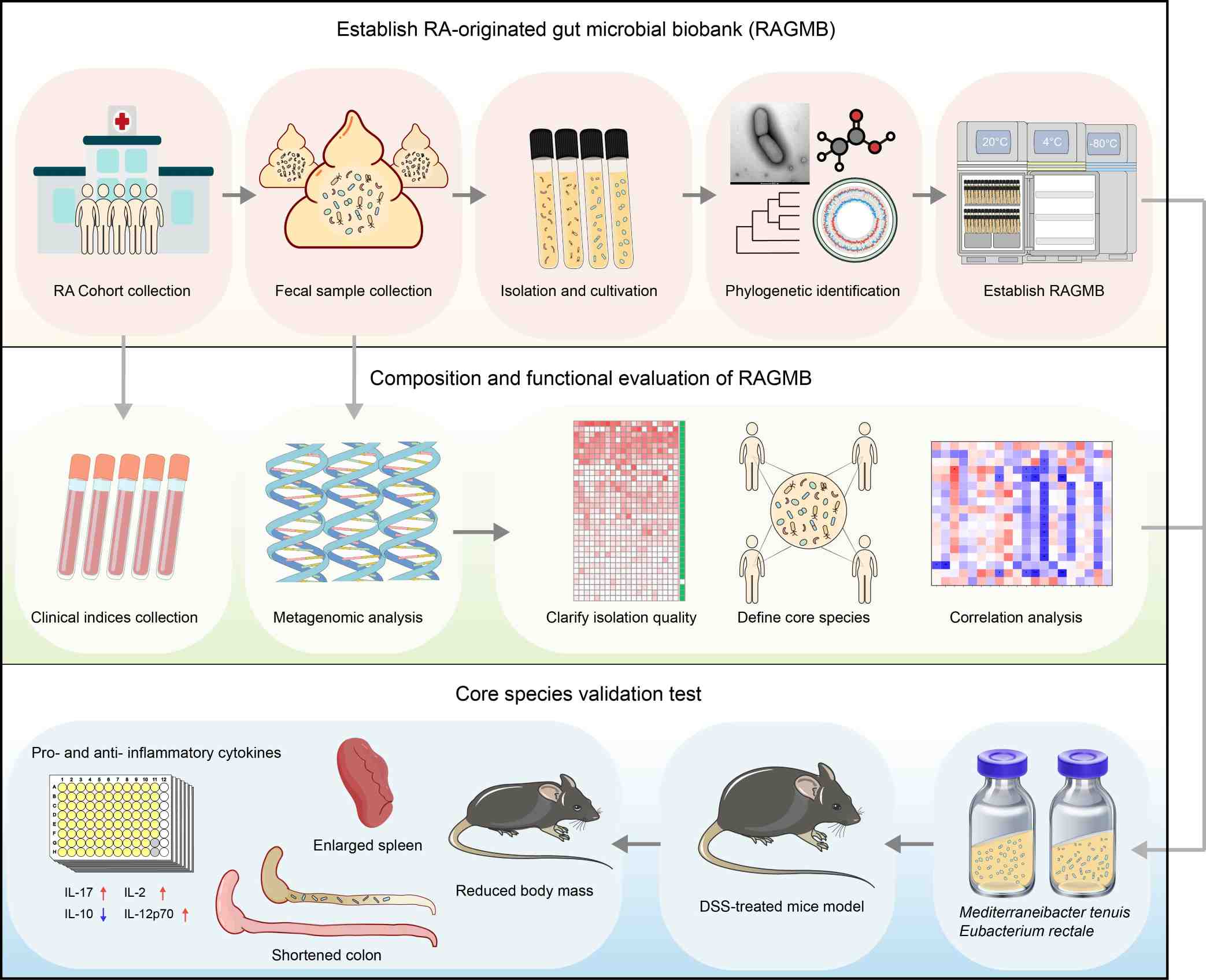
Intensive collection of 3200 bacterial isolates from fecal samples of newly diagnosed rheumatoid arthritis (RA) patients resulted in an RA-originated gut microbial biobank (RAGMB). This RAGMB has 601 strains that represent 280 bacterial species (including 43 novel species), and covers 93.2% of medium- and high-abundant gut microbial species of the RA fecal samples. By integrating additional RA cohort metagenomic data, an RA core microbiome composing of 20 core microbial species were defined and correlated to RA clinical indices. Two RA core microbial species, Mediterraneibacter tenuis and Eubacterium rectale, were selected for experimental validation with mouse models and the results showed that both M. tenuis and E. rectale exacerbated host inflammatory and immune responses.
LTBR acts as a novel immune checkpoint of tumor-associated macrophages for cancer immunotherapy
- 04 September 2024
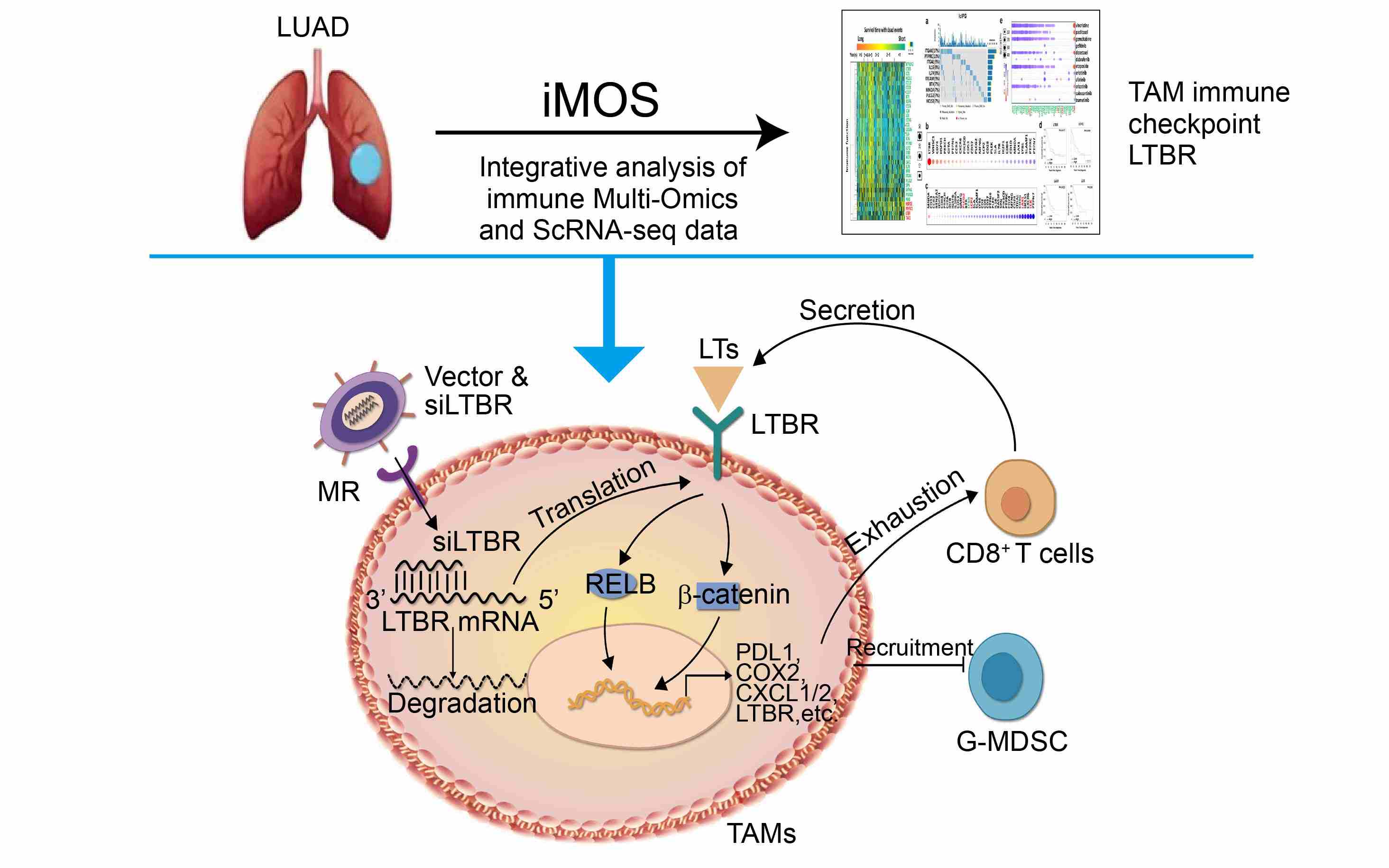
Through integrative analysis of immune multiomics data and single-cell RNA-seq data, this study identifies lymphotoxin β receptor (LTBR) as a potential immune checkpoint of tumor-associated macrophages (TAMs). LTBR+ TAMs are associated with lung adenocarcinoma stages, immunotherapy failure, and poor prognosis. Mechanistically, LTΒR maintains TAM immunosuppressive activity and M2 phenotype by noncanonical nuclear factor kappa B and Wnt/β-catenin signaling pathways. Disruption of LTΒR in TAMs enhances the therapeutic effect of cancer immunotherapy.
tigeR: Tumor immunotherapy gene expression data analysis R package
- 06 August 2024
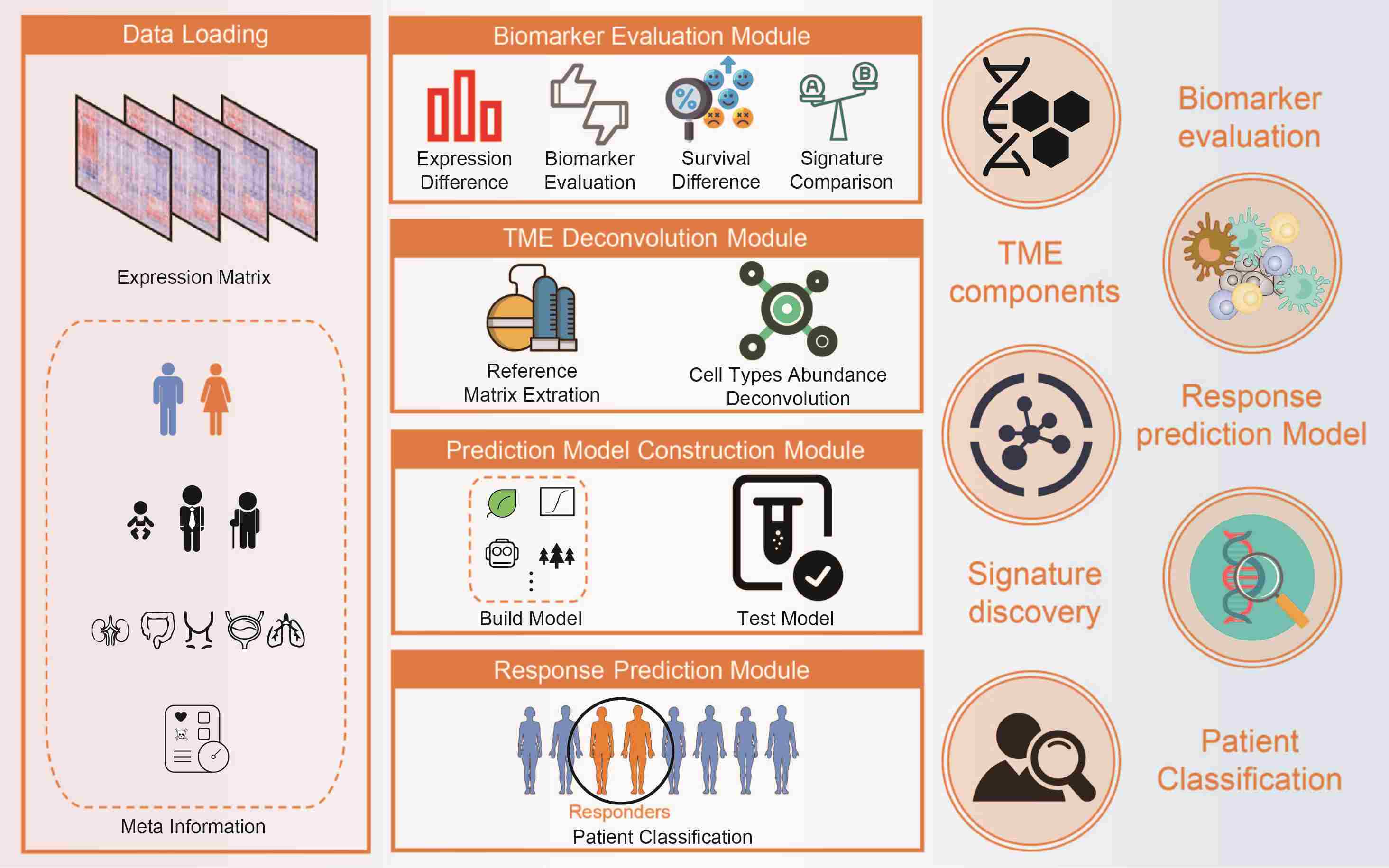
The Tumor Immunotherapy Gene Expression R package (tigeR) toolkit provides four distinct yet closely interconnected modules, including the Biomarker Evaluation module, Tumor Microenvironment Deconvolution module, Prediction Model Construction module, and Response Prediction module, to explore biomarkers and construct predictive models via built-in or custom immunotherapy gene expression data. With a comprehensive suite of functionalities, tigeR not only streamlines the analysis process but also catalyzes discoveries in the realm of tumor immunotherapy.
Integrating genome- and transcriptome-wide association studies to uncover the host–microbiome interactions in bovine rumen methanogenesis
- 03 September 2024
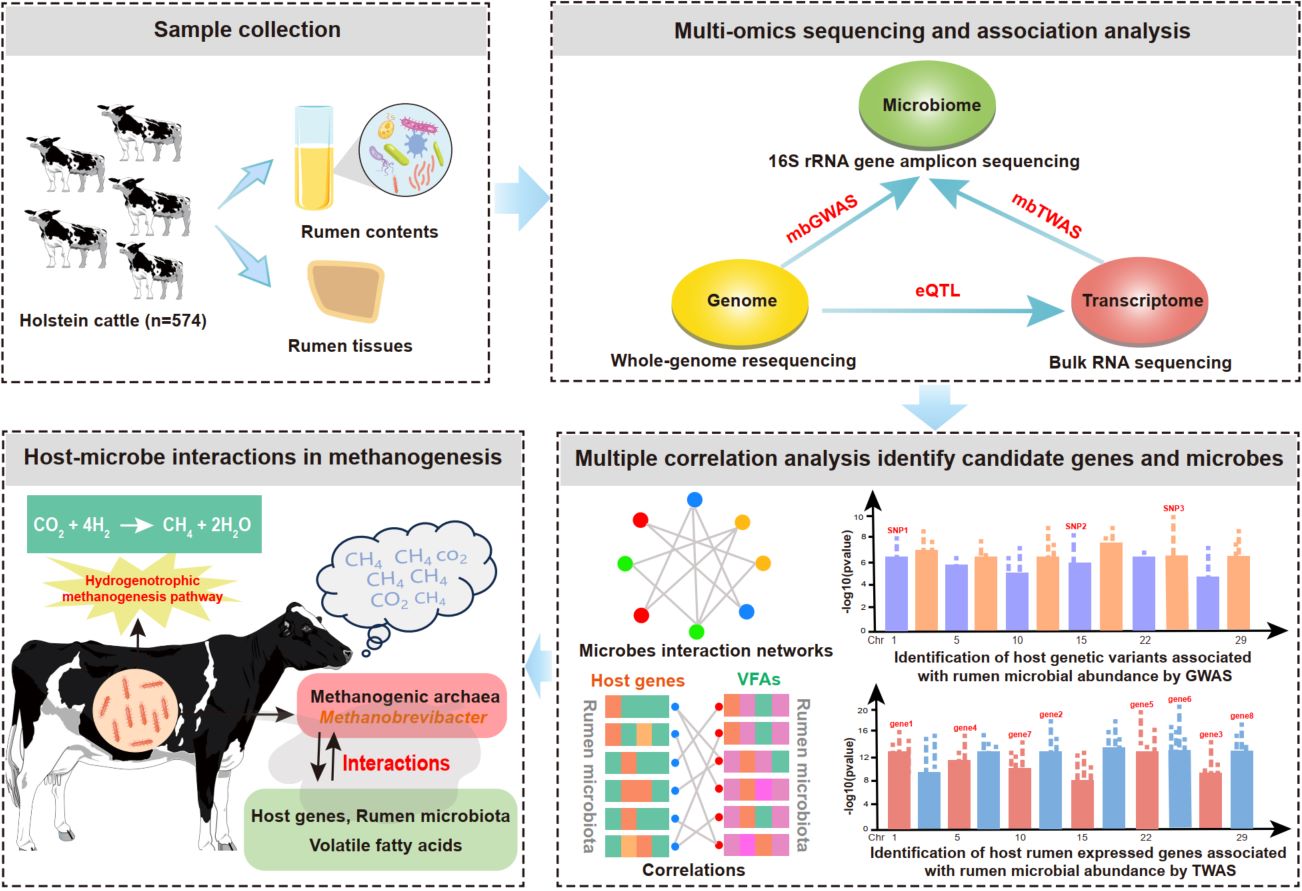
We systematically evaluated the relationship between host and microorganisms using matched host genome, rumen transcriptome, and microbiome data from a cohort of 574 Holstein cattle. The associations between rumen gene expression and rumen microbiota abundance via transcriptome-wide association study (TWAS) may provide more molecular markers at the gene level, which bridge the host–microbe interactions. Methanogens belonging to Methanobrevibacter genus are common hydrogenotrophic methanogenic archaea, which usually utilize hydrogen (H2) and carbon dioxide (CO2) produced from microbial fermentation as substrates to produce methane (CH4). By combining multiple correlation analyses based on host TWAS genes, rumen microbiota, and volatile fatty acids, we illustrated that host–microbe interactions in the hydrogenotrophic methanogenesis pathway are possibly involved in substrate hydrogen metabolism and transport. This study broadens our insights into the potential mechanisms of host–microbe interactions in rumen methanogenesis.
TCellSI: A novel method for T cell state assessment and its applications in immune environment prediction
- 26 August 2024
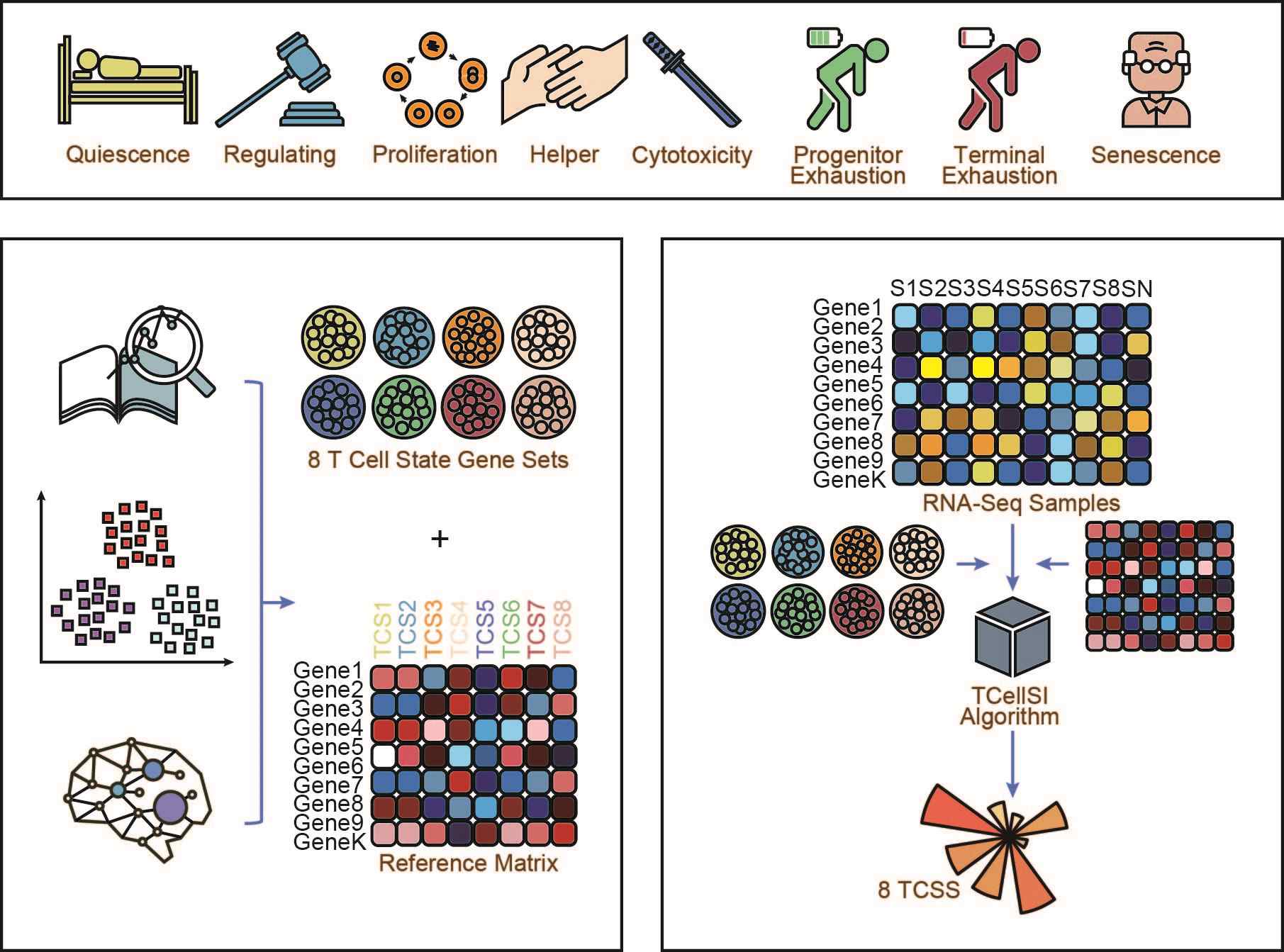
TCellSI is a novel method to evaluate T cell states via transcriptome data, using specific marker gene sets and a compiled reference spectrum. TCellSI calculates T cell state scores for eight states: quiescence, regulating, proliferation, helper, cytotoxicity, progenitor exhaustion, terminal exhaustion, and senescence, offering valuable insights into the immune environment.
Sangerbox 2: Enhanced functionalities and update for a comprehensive clinical bioinformatics data analysis platform
- 02 September 2024
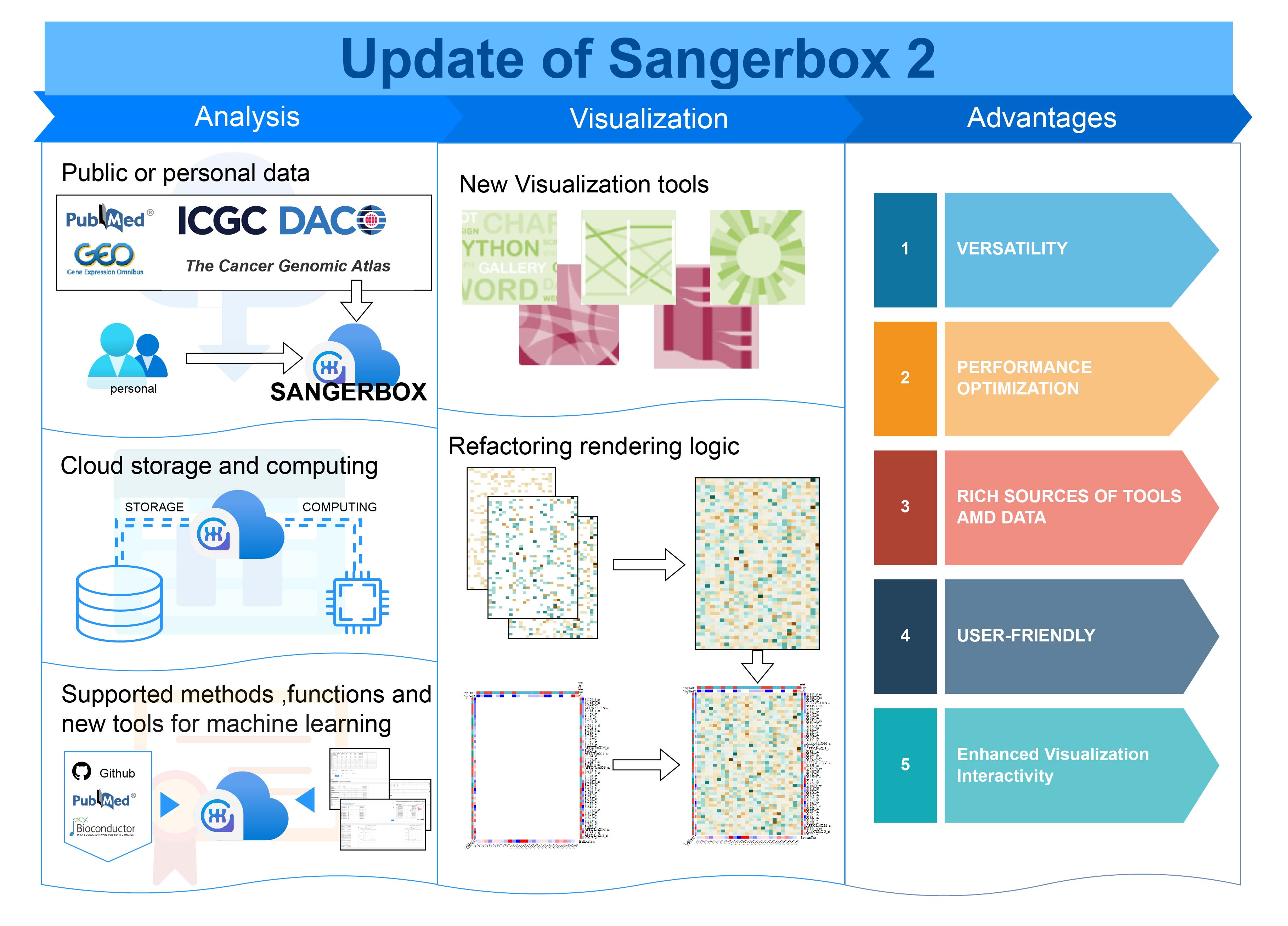
This abstract showcases the key enhancements of SangerBox 2, focusing on versatility, performance optimization, rich visualization tools, user-friendliness, and enhanced interactivity. The platform integrates advanced machine learning tools, improves computational efficiency, and introduces new visualization options like wordclouds and Manhattan plots. Interactive features allow real-time heatmap adjustments, greatly enhancing the user experience. SangerBox 2 supports both public and personal data, utilizing cloud storage and computing, making it a versatile tool for bioinformatics research.
Novel microbial modifications of bile acids and their functional implications
- 03 October 2024
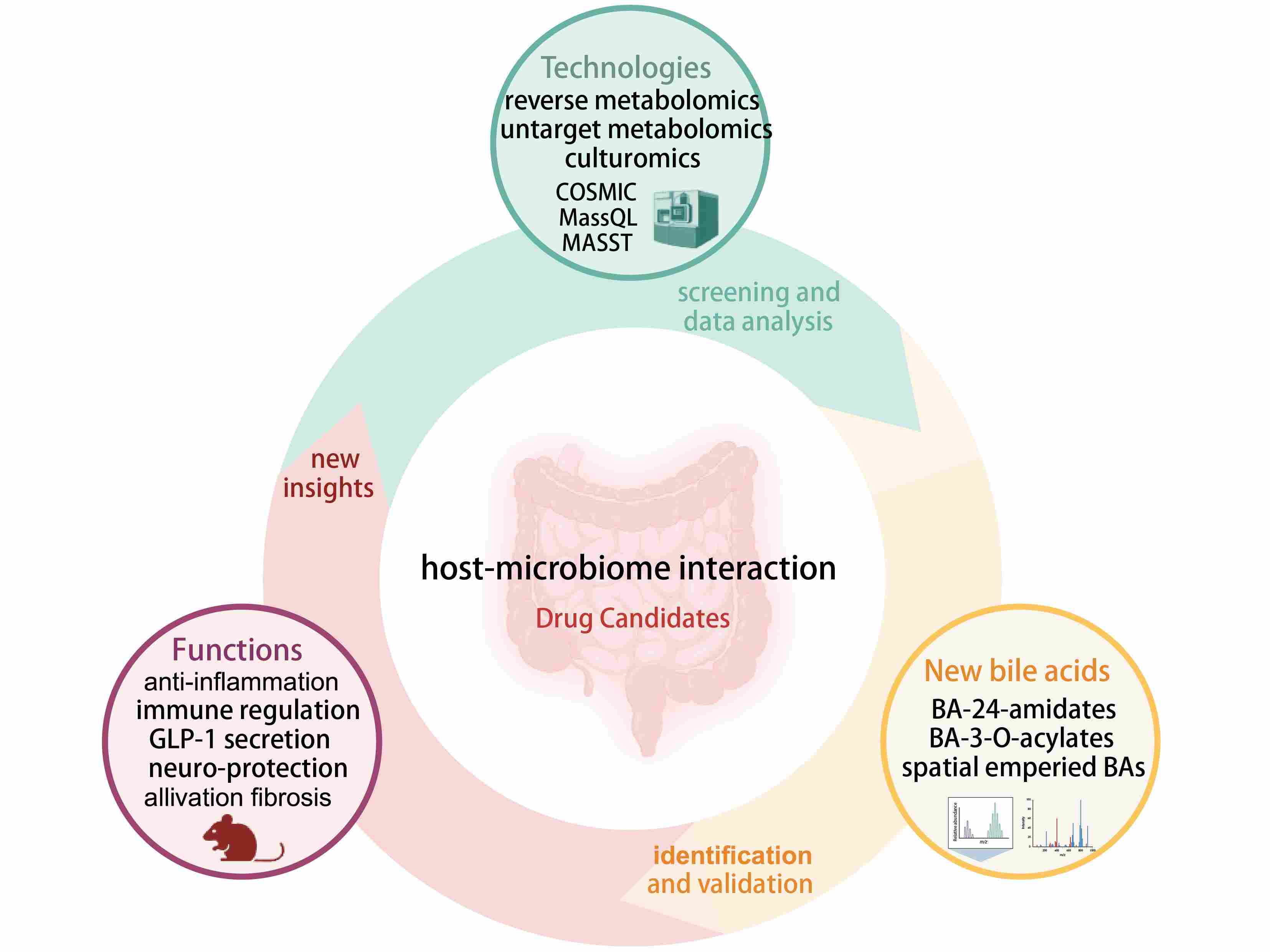
This review outlines the recent discoveries of bile acids that have undergone novel microbial modifications, highlighting their biological roles and the profound implications for the development of innovative therapeutic strategies. The review aims to provide valuable insights and breakthroughs for future drug candidates in the expanding field of bile acid therapeutics.
Multi-level insights into the immuno-oncology-microbiome axis: From biotechnology to novel therapies
- 07 September 2024
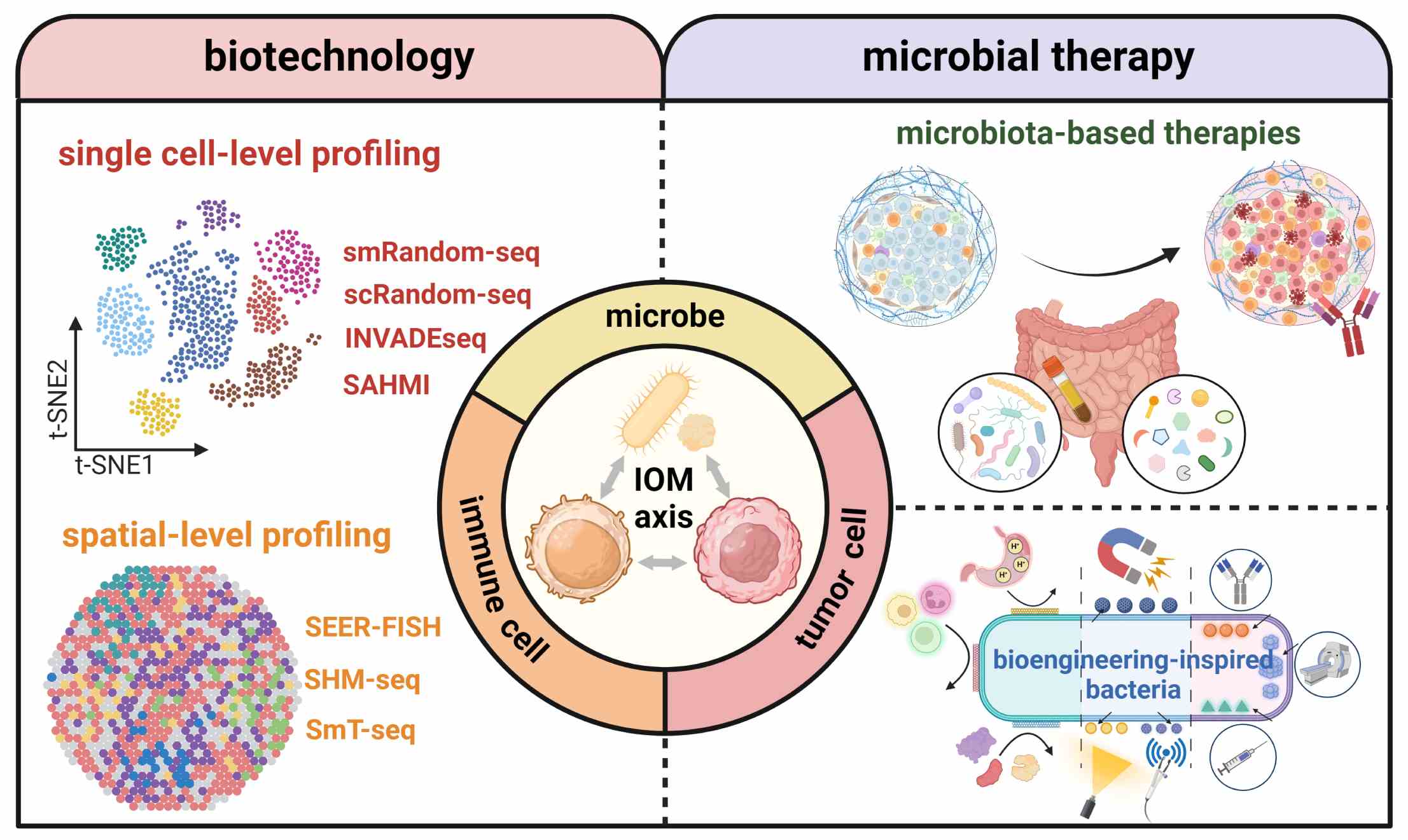
The multifaceted interactions among the immune system, cancer cells and microbial components have established a novel concept of the immuno-oncology-microbiome (IOM) axis. Microbiome sequencing technologies have played a pivotal role in not only analyzing how gut microbiota affect local and distant tumors, but also providing unprecedented insights into the intratumor host-microbe interactions. Herein, we discuss the emerging trends of transiting from bulk-level to single cell- and spatial-level analyses. Moving forward with advances in biotechnology, microbial therapies, including microbiota-based therapies and bioengineering-inspired microbes, will add diversity to the current oncotherapy paradigm.
Efficient and easy-to-use capturing three-dimensional metagenome interactions with GutHi-C
- 22 July 2024
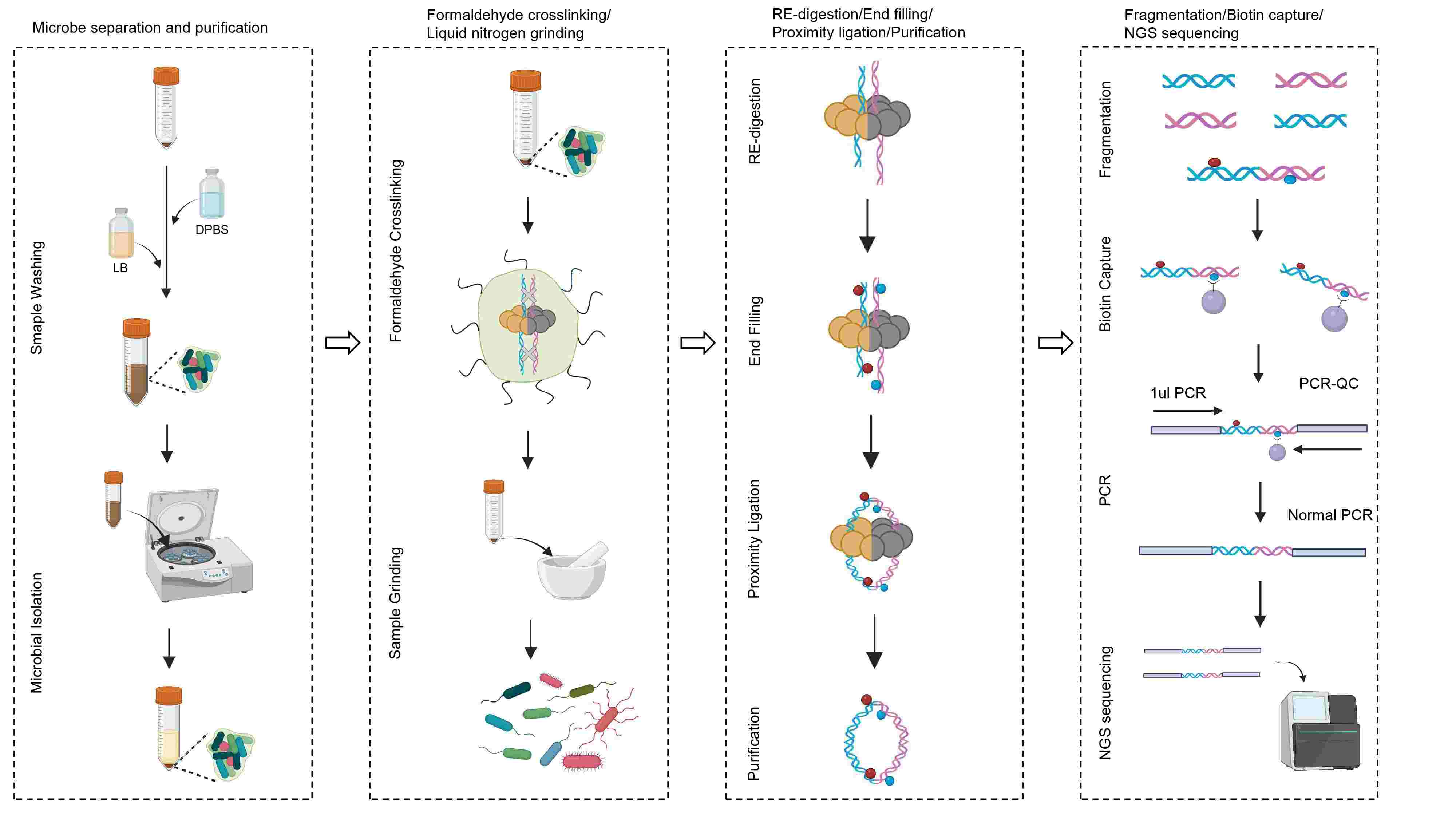
Hi-C can obtain three-dimensional chromatin structure information and is widely used for genome assembly. We constructed the GutHi-C technology. As shown in the graphical abstract, it is a highly efficient and quick-to-operate method and can be widely used for human, livestock, and poultry gut microorganisms. It provides a reference for the Hi-C methodology of the microbial metagenome. DPBS, Dulbecco's phosphate-buffered saline; Hi-C, high-through chromatin conformation capture; LB, Luria-Bertani; NGS, next-generation sequencing; PCR, polymerase chain reaction; QC, quality control.
Role of respiratory system microbiota in development of lung cancer and clinical application
- 25 August 2024
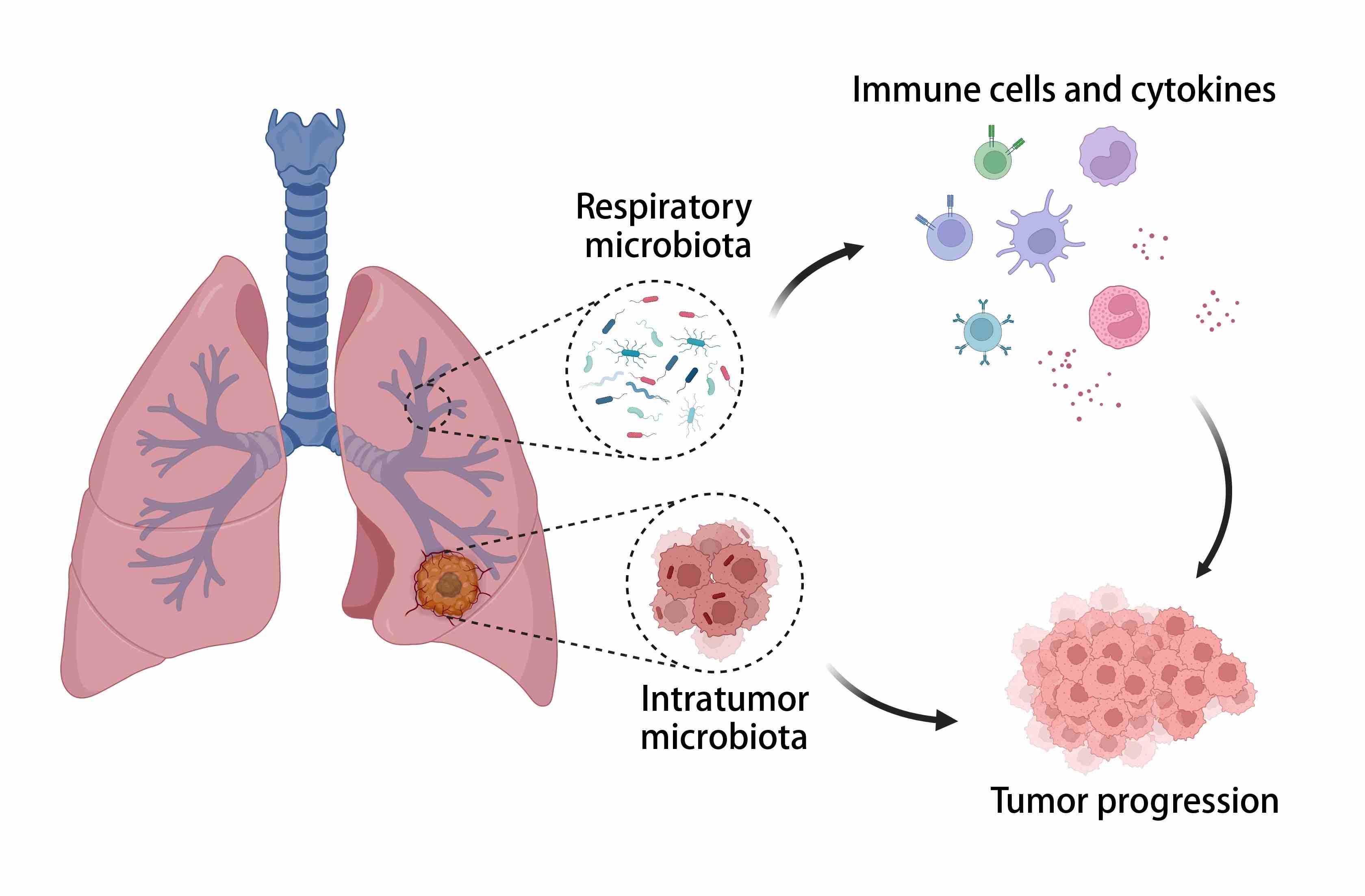
Microbes play a significant role in human tumor development and profoundly impact treatment efficacy, particularly in immunotherapy. The respiratory tract extensively interacts with the external environment and possesses a mucosal immune system. This prompts consideration of the relationship between respiratory microbiota and lung cancer. Advancements in culture-independent techniques have revealed unique communities within the lower respiratory tract. Here, we provide an overview of the respiratory microbiota composition, dysbiosis characteristics in lung cancer patients, and microbiota profiles within lung cancer. We delve into how the lung microbiota contributes to lung cancer onset and progression through direct functions, sustained immune activation, and immunosuppressive mechanisms. Furthermore, we emphasize the clinical utility of respiratory microbiota in prognosis and treatment optimization for lung cancer.
Dietary therapies interlinking with gut microbes toward human health: Past, present, and future
- 10 August 2024
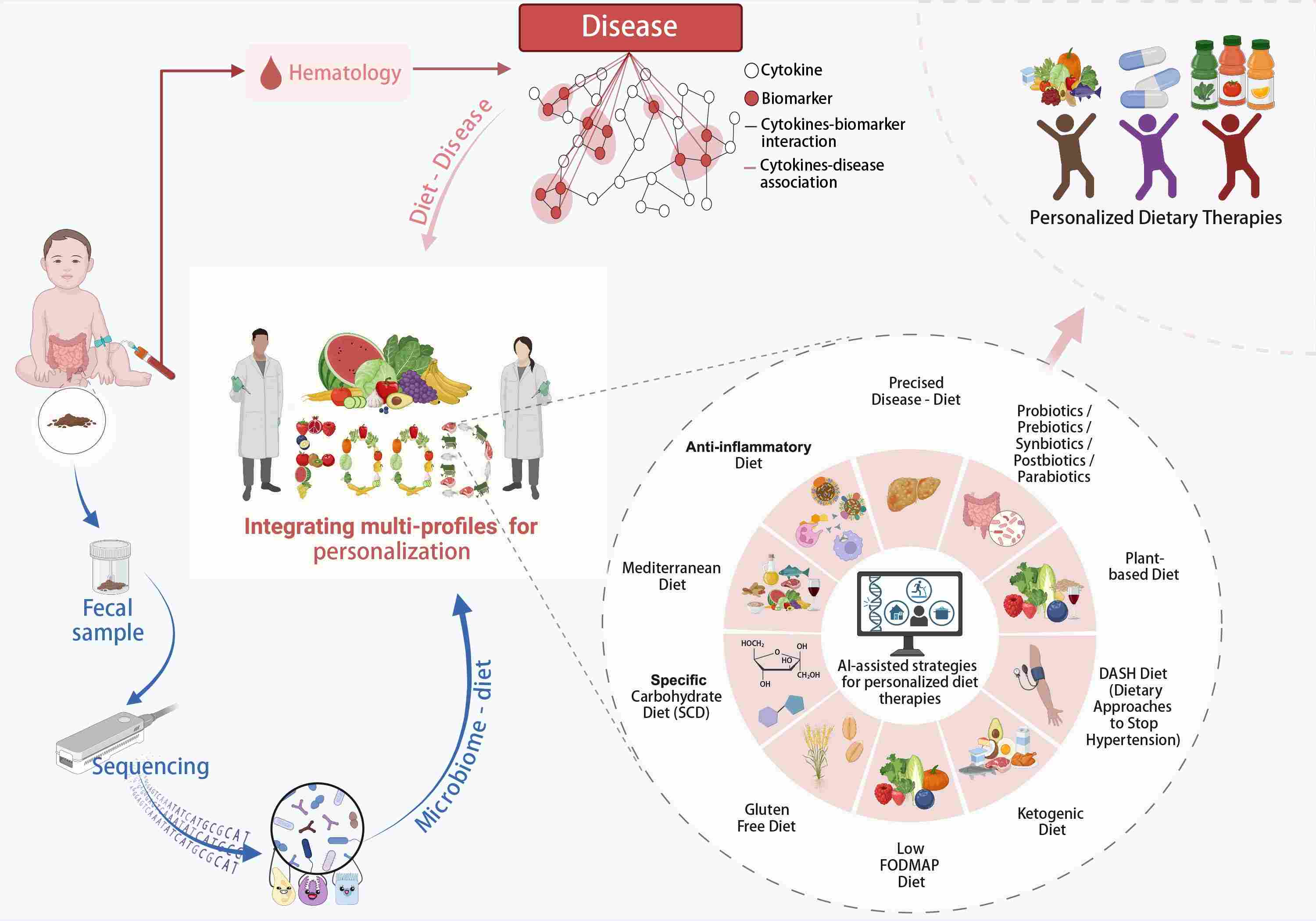
Overview of personalized dietary therapies. This flow chart exhibits the future prospect for integrating human microbiome and bio-medical research to revolutionize the precise personalized dietary therapies. With the development of artificial intelligence (AI), incorporating database may achieve personalized dietary therapies with high precision.
OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization
- 01 August 2024
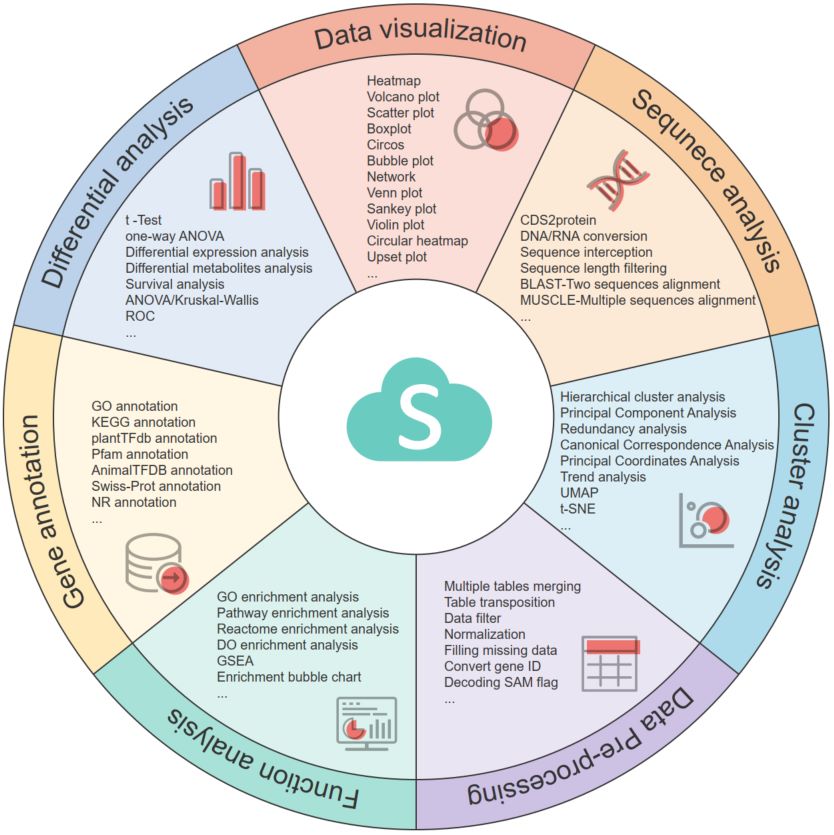
The OmicShare tools platform is a user-friendly online resource for data analysis and visualization, encompassing 161 bioinformatic tools. Users can easily track the progress of projects in real-time through an overview interface. The platform has a powerful interactive graphics engine that allows for the custom-tailored modification of charts generated from analyses. The visually appealing charts produced by OmicShare improve data interpretability and meet the requirements for publication. It has been acknowledged in over 4000 publications and is available in https://www.omicshare.com/tools/.
USEARCH 12: Open-source software for sequencing analysis in bioinformatics and microbiome
- 02 September 2024
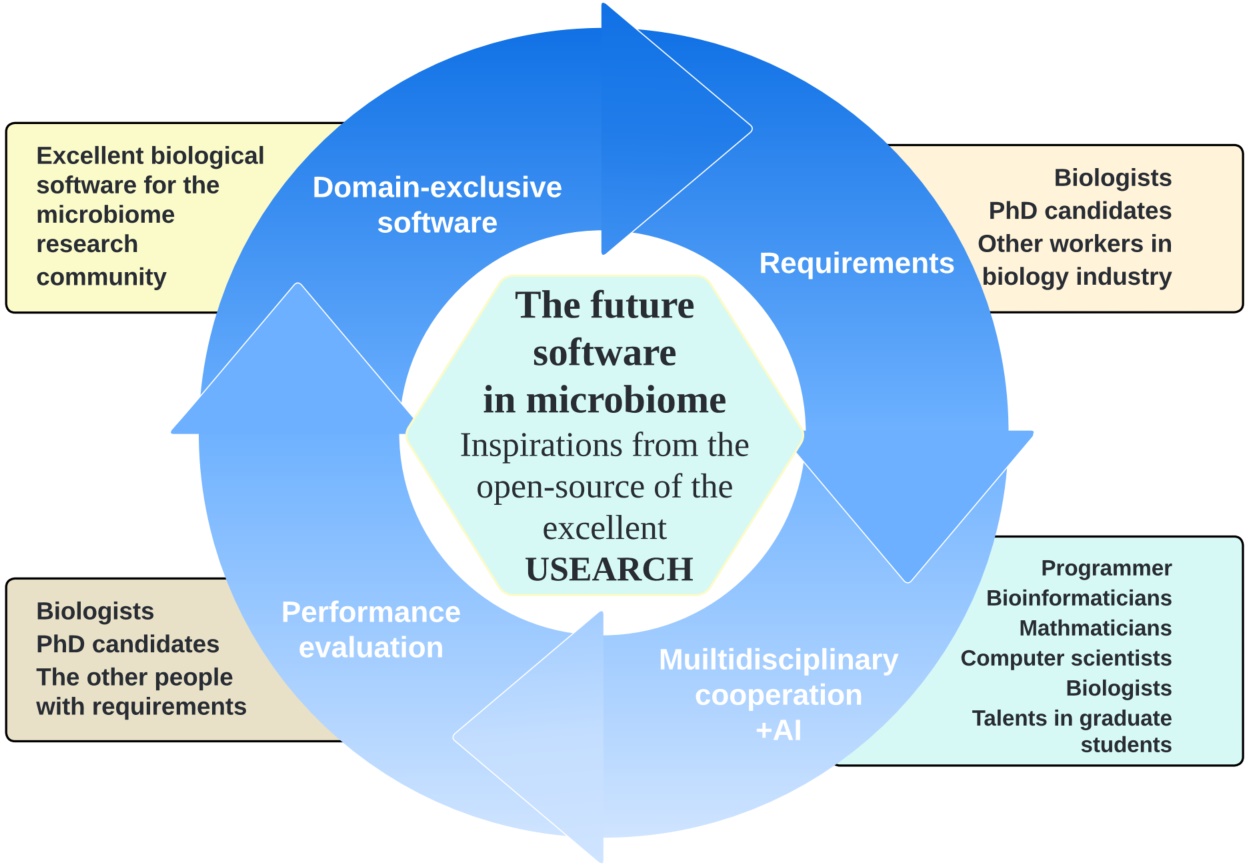
The well-known bioinformatic software USEARCH v12 was open sourced. Its meaning encourages the microbiome research community to constantly develop excellent bioinformatic software based on the codes. The open source and popularization of artificial intelligence (AI) will make a better infrastructure for microbiome research.
Akkermansia muciniphila administration ameliorates streptozotocin-induced hyperglycemia and muscle atrophy by promoting IGF2 secretion from mouse intestine
- 01 October 2024
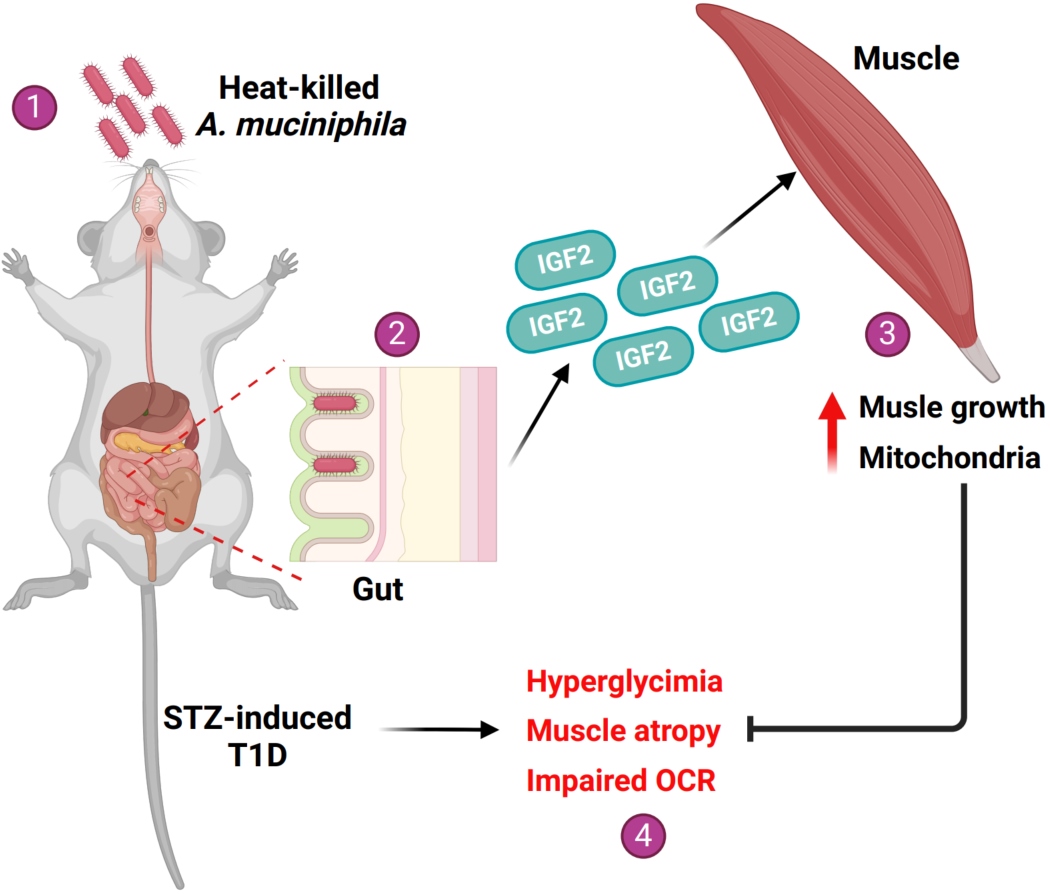
Type 1 diabetes mellitus (T1DM) is an autoimmune disease that can lead to severe diabetic complications. While the changes and correlations between gut microbiota and the pathogenesis of T1DM have been extensively studied, little is known about the benefits of interventions on gut bacterial communities, particularly using probiotics, for this disease. In the present study, we reported that the mice surviving after 5 months of streptozotocin (STZ) injection had reduced blood glucose level and recovered gut microbiota with increased Akkermansia muciniphila proportion. Gavage of heat-killed A. muciniphila increases the diversity of gut microbiota and elevated immune and metabolic signaling pathways in the intestine. Mechanistically, A. muciniphila treatment promoted the secretion of insulin-like growth factor 2 (IGF2) which subsequently activated IGF2 signaling in skeletal muscles and enhanced muscle and global metabolism. Our results suggest that the administration of heat-killed A. muciniphila could be a potential therapeutic strategy for T1DM and its associated hyperglycemia.